This is a read-only mirror of pymolwiki.org
Difference between revisions of "Main Page"
Jump to navigation
Jump to search
(Added link to LigAlign.) |
m (Modified drawMinBoundingBox) |
||
| (17 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
|- | |- | ||
|} | |} | ||
| − | |||
| − | |||
{| align="center" style="width:90%; margin-bottom: 20px" | {| align="center" style="width:90%; margin-bottom: 20px" | ||
| | | | ||
| Line 40: | Line 38: | ||
| | | | ||
|} | |} | ||
| − | |||
{| align="center" width="90%" style="background: #fafafa; border-right: 1px solid #333; border-left: 1px solid #333; border-bottom: 1px solid #333" | {| align="center" width="90%" style="background: #fafafa; border-right: 1px solid #333; border-left: 1px solid #333; border-bottom: 1px solid #333" | ||
|+ style="text-align: left; font-weight:bold; font-size:150%; color:#333; background: #EFE6FF; padding:10px; border: 1px solid #333" | News and Updates ([[Older_News|archive]]) | |+ style="text-align: left; font-weight:bold; font-size:150%; color:#333; background: #EFE6FF; padding:10px; border: 1px solid #333" | News and Updates ([[Older_News|archive]]) | ||
| Line 51: | Line 48: | ||
! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Wiki | ! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Wiki | ||
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ Documented a few more settings. Also, check out the cool, [[Huge_surfaces]] page for handling very large objects and representing them as surfaces, in PyMOL. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ We got our '''2,000,000th''' page view! |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ Added links in the table on the Main Page (above) for submitting & tracking bugs and feature reqeuests. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ New logo for the wiki. It's DNA. You can easily see the major/minor grooves. If you don't see it, force a reload of the page (CTRL-F5, usually). |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ New category about PyMOL [[:Category:Performance|performance]]: making the impossible possible, and the difficult easier/faster. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | |valign="bottom" width="150px" style="padding: 0 20px 20px 0; clear:right;" |[[Image: | + | |valign="bottom" width="150px" style="padding: 0 20px 20px 0; clear:right;" |[[Image:080701_h.a.steinberg_biochemie.jpg|125px]] Sample Cover from the [[Covers]] gallery. |
|} | |} | ||
|- | |- | ||
| Line 79: | Line 68: | ||
|- | |- | ||
! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | PyMOL | ! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | PyMOL | ||
| + | |- | ||
| + | | style="padding: 3px 5px 10px 15px;"| ♦ '''PyMOL now integrates seamlessly with MS Powerpoint. See [[Axpymol]].''' | ||
|- | |- | ||
| style="padding: 3px 5px 10px 15px;"| ♦ PyMOL now comes with some builtin examples: look in the '''examples''' directory of your source tree. | | style="padding: 3px 5px 10px 15px;"| ♦ PyMOL now comes with some builtin examples: look in the '''examples''' directory of your source tree. | ||
| Line 88: | Line 79: | ||
| style="padding: 3px 5px 10px 15px;"| ♦ [[Ellipsoids]] representation added for drawing thermal ellipsoids. | | style="padding: 3px 5px 10px 15px;"| ♦ [[Ellipsoids]] representation added for drawing thermal ellipsoids. | ||
|} | |} | ||
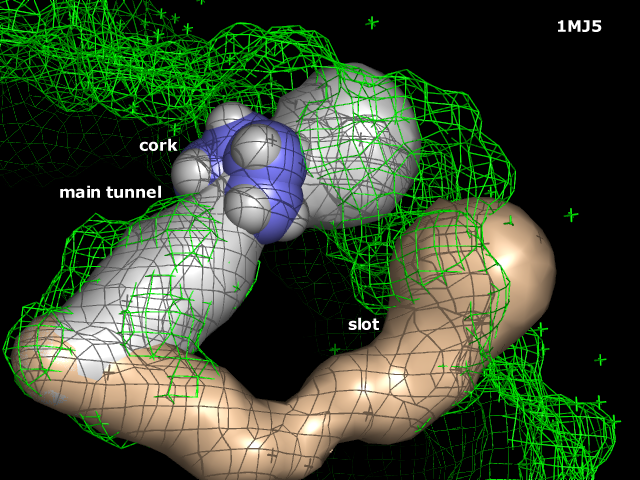
| − | |valign="bottom" width="150px" style="padding: 0 20px 20px 0" |[[Image: | + | |valign="bottom" width="150px" style="padding: 0 20px 20px 0" |[[Image:Caver.png|125px]] Screenshot of [[Caver]] showing cavities. |
|} | |} | ||
|- | |- | ||
| Line 98: | Line 89: | ||
! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Scripts & Plugins | ! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Scripts & Plugins | ||
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ [[GetNamesInSel]] — find the names of all objects in a selection. |
| + | |- | ||
| + | | style="padding: 3px 5px 10px 15px;"| ♦ [[CalcArea]] — find the area of any given object/selection; | ||
| + | |- | ||
| + | | style="padding: 3px 5px 10px 15px;"| ♦ [[ConnectedCloud]] — find connected clouds of objects in PyMOL. | ||
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ | + | | style="padding: 3px 5px 10px 15px;"| ♦ [[MakeVinaCommand]] — Use PyMOL to create a valid command line for the new [http://vina.scripps.edu Vina] docking software. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [[ | + | | style="padding: 3px 5px 10px 15px;"| ♦ [[DrawBoundingBox]] — Draw a bounding box around your selection. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [[ | + | | style="padding: 3px 5px 10px 15px;"| ♦ Locate loaded PyMOL objects that are nearby some other object with [[FindObjectsNearby]]. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [ | + | | style="padding: 3px 5px 10px 15px;"| ♦ Ever wanted to load all the PDBs in a directory within PyMOL? Now you can load all files in a dir with [[LoadDir]]. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [[ | + | | style="padding: 3px 5px 10px 15px;"| ♦ [[PDB Web Services Script]] — Example using PyMOL and the PDB Web Services. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [[ | + | | style="padding: 3px 5px 10px 15px;"| ♦ [[LigAlign]] — Ligand-based active site alignment and comparison. |
|- | |- | ||
| − | | style="padding: 3px 5px 10px 15px;"| ♦ [[ | + | | style="padding: 3px 5px 10px 15px;"| ♦ Added a [[COM|simple script]] for finding the center or mass, or moving a selection to the origin. |
|} | |} | ||
| − | |width="150px" style="padding: 0 20px 20px 0; text-align:left" |[[Image: | + | |width="150px" style="padding: 0 20px 20px 0; text-align:left" |[[Image:DrawMinBB.png|165px]] Screenshot of [[DrawBoundingBox]] in action. |
|} | |} | ||
|} | |} | ||
Revision as of 19:11, 30 March 2009
| We are the community-based support site for the popular molecular visualization program, PyMOL. |
|
|||||||||||||||||||
| |||||||||||||
| |||||||||||||
|