This is a read-only mirror of pymolwiki.org
Difference between revisions of "BbPlane"
Jump to navigation
Jump to search
(support multiple chains and proline) |
(improve speed, support two_sided_lighting) |
||
| Line 23: | Line 23: | ||
# | # | ||
# Author: Jason Vertrees, 06/2010 | # Author: Jason Vertrees, 06/2010 | ||
| + | # Modified by Thomas Holder, 06/2010 | ||
# Copyright (C) Schrodinger | # Copyright (C) Schrodinger | ||
# Open Source License: MIT | # Open Source License: MIT | ||
| Line 32: | Line 33: | ||
def bbPlane(objSel='(all)', color='white', transp=0.0): | def bbPlane(objSel='(all)', color='white', transp=0.0): | ||
""" | """ | ||
| + | DESCRIPTION | ||
| + | |||
Draws a plane across the backbone for a selection | Draws a plane across the backbone for a selection | ||
| − | + | ARGUMENTS | |
| − | objSel | + | |
| − | + | objSel = string: protein object or selection {default: (all)} | |
color = string: color name or number {default: white} | color = string: color name or number {default: white} | ||
| Line 42: | Line 45: | ||
transp = float: transparency component (0.0--1.0) {default: 0.0} | transp = float: transparency component (0.0--1.0) {default: 0.0} | ||
| − | + | NOTES | |
| − | |||
| − | + | You need to pass in an object or selection with at least two | |
| − | |||
amino acids. The plane spans CA_i, O_i, N-H_(i+1), and CA_(i+1) | amino acids. The plane spans CA_i, O_i, N-H_(i+1), and CA_(i+1) | ||
| − | |||
| − | |||
| − | |||
""" | """ | ||
# format input | # format input | ||
transp = float(transp) | transp = float(transp) | ||
stored.AAs = [] | stored.AAs = [] | ||
| + | coords = dict() | ||
| − | # need hydrogens | + | # need hydrogens on peptide nitrogen |
| − | cmd.h_add(objSel) | + | cmd.h_add('(%s) and n. N' % objSel) |
# get the list of residue ids | # get the list of residue ids | ||
for obj in cmd.get_object_list(objSel): | for obj in cmd.get_object_list(objSel): | ||
| − | + | sel = obj + " and (" + objSel + ")" | |
| + | for a in cmd.get_model(sel + " and n. CA").atom: | ||
| + | key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi) | ||
| + | stored.AAs.append(key) | ||
| + | coords[key] = [a.coord,None,None] | ||
| + | for a in cmd.get_model(sel + " and n. O").atom: | ||
| + | key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi) | ||
| + | if key in coords: | ||
| + | coords[key][1] = a.coord | ||
| + | for a in cmd.get_model(sel + " and ((n. N extend 1 and e. H) or (r. PRO and n. CD))").atom: | ||
| + | key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi) | ||
| + | if key in coords: | ||
| + | coords[key][2] = a.coord | ||
# need at least two amino acids | # need at least two amino acids | ||
| Line 78: | Line 89: | ||
curIdx, nextIdx = str(stored.AAs[res]), str(stored.AAs[res+1]) | curIdx, nextIdx = str(stored.AAs[res]), str(stored.AAs[res+1]) | ||
| − | + | # populate the position array | |
| − | + | pos = [coords[curIdx][0], coords[curIdx][1], coords[nextIdx][2], coords[nextIdx][0]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
# if the data are incomplete for any residues, ignore | # if the data are incomplete for any residues, ignore | ||
| − | if | + | if None in pos: |
print 'peptide bond %s -> %s incomplete' % (curIdx, nextIdx) | print 'peptide bond %s -> %s incomplete' % (curIdx, nextIdx) | ||
continue | continue | ||
| − | |||
| − | |||
| − | |||
if cpv.distance(pos[0], pos[3]) > 4.0: | if cpv.distance(pos[0], pos[3]) > 4.0: | ||
| Line 102: | Line 102: | ||
# fill in the vertex data for the triangles; | # fill in the vertex data for the triangles; | ||
| − | + | for i in [0,1,2,3,2,1]: | |
| − | + | obj.append(VERTEX) | |
| − | + | obj.extend(pos[i]) | |
| − | |||
| − | |||
| − | |||
| − | VERTEX | ||
| − | |||
| − | |||
| − | |||
# finish the CGO | # finish the CGO | ||
Revision as of 11:52, 30 June 2010
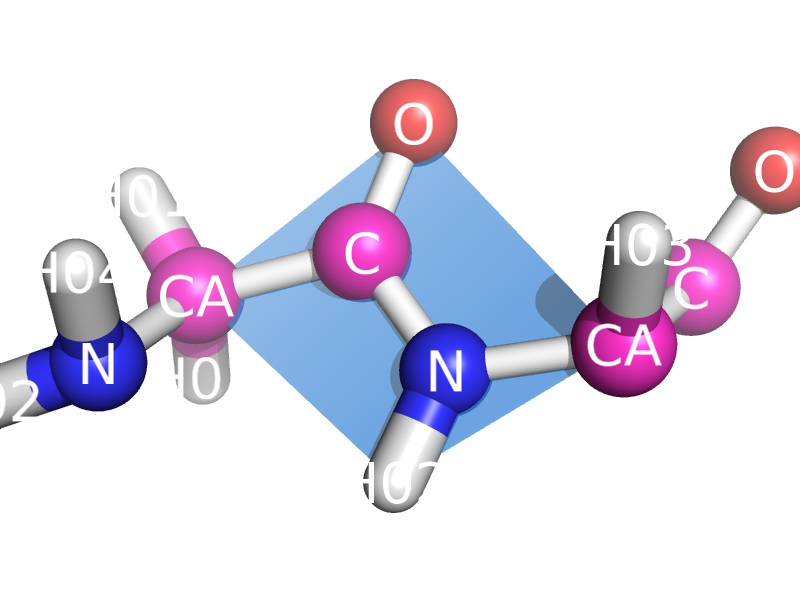
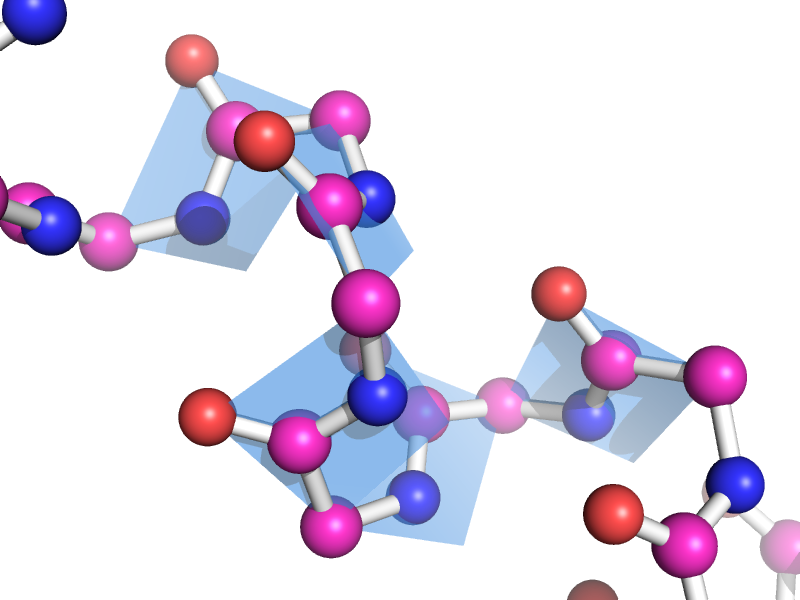
This script will draw a CGO plane between the backbone atoms of two neighboring residues. This is to show the planarity of the atoms. The image style this is meant to represent can be found many places, like "Introduction to Protein Structure" by Branden and Tooze (2nd ed. pp. 8).
Examples
# download the source and save as bbPlane.py
run bbPlane.py
fetch 1cll
# make planes for residues 4-9
bbPlane i. 4-10
The Source
#
# -- bbPLane.py - draws a CGO plane across the backbone atoms of
# neighboring amino acids
#
# Author: Jason Vertrees, 06/2010
# Modified by Thomas Holder, 06/2010
# Copyright (C) Schrodinger
# Open Source License: MIT
#
from pymol.cgo import * # get constants
from pymol import cmd, stored
from chempy import cpv
def bbPlane(objSel='(all)', color='white', transp=0.0):
"""
DESCRIPTION
Draws a plane across the backbone for a selection
ARGUMENTS
objSel = string: protein object or selection {default: (all)}
color = string: color name or number {default: white}
transp = float: transparency component (0.0--1.0) {default: 0.0}
NOTES
You need to pass in an object or selection with at least two
amino acids. The plane spans CA_i, O_i, N-H_(i+1), and CA_(i+1)
"""
# format input
transp = float(transp)
stored.AAs = []
coords = dict()
# need hydrogens on peptide nitrogen
cmd.h_add('(%s) and n. N' % objSel)
# get the list of residue ids
for obj in cmd.get_object_list(objSel):
sel = obj + " and (" + objSel + ")"
for a in cmd.get_model(sel + " and n. CA").atom:
key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi)
stored.AAs.append(key)
coords[key] = [a.coord,None,None]
for a in cmd.get_model(sel + " and n. O").atom:
key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi)
if key in coords:
coords[key][1] = a.coord
for a in cmd.get_model(sel + " and ((n. N extend 1 and e. H) or (r. PRO and n. CD))").atom:
key = '/%s/%s/%s/%s' % (obj,a.segi,a.chain,a.resi)
if key in coords:
coords[key][2] = a.coord
# need at least two amino acids
if len(stored.AAs) <= 1:
print "ERROR: Please provide at least two amino acids, the alpha-carbon on the 2nd is needed."
return
# prepare the cgo
obj = [
BEGIN, TRIANGLES,
COLOR,
]
obj.extend(cmd.get_color_tuple(color))
for res in range(0, len(stored.AAs)-1):
curIdx, nextIdx = str(stored.AAs[res]), str(stored.AAs[res+1])
# populate the position array
pos = [coords[curIdx][0], coords[curIdx][1], coords[nextIdx][2], coords[nextIdx][0]]
# if the data are incomplete for any residues, ignore
if None in pos:
print 'peptide bond %s -> %s incomplete' % (curIdx, nextIdx)
continue
if cpv.distance(pos[0], pos[3]) > 4.0:
print '%s and %s not adjacent' % (curIdx, nextIdx)
continue
# fill in the vertex data for the triangles;
for i in [0,1,2,3,2,1]:
obj.append(VERTEX)
obj.extend(pos[i])
# finish the CGO
obj.append(END)
# update the UI
newName = cmd.get_unused_name("backbonePlane")
cmd.load_cgo(obj, newName)
cmd.set("cgo_transparency", transp, newName)
cmd.extend("bbPlane", bbPlane)