This is a read-only mirror of pymolwiki.org
Difference between revisions of "Align"
m |
m (1 revision) |
||
| (34 intermediate revisions by 12 users not shown) | |||
| Line 1: | Line 1: | ||
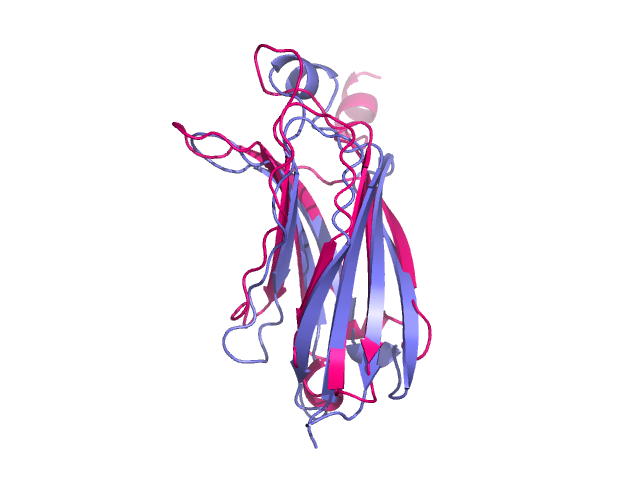
| − | + | [[Image:after_alignment.png|400px|thumb|right|Two proteins after structure alignment]] | |
| − | |||
| − | |||
| − | |||
| − | === | + | '''align''' performs a sequence alignment followed by a structural superposition, and then carries out zero or more cycles of refinement in order to reject structural outliers found during the fit. |
| + | [[align]] does a good job on proteins with decent sequence similarity (identity >30%). | ||
| + | For comparing proteins with lower sequence identity, the [[super]] and [[cealign]] commands perform better. | ||
| + | |||
| + | == Usage == | ||
| + | |||
| + | align mobile, target [, cutoff [, cycles | ||
| + | [, gap [, extend [, max_gap [, object | ||
| + | [, matrix [, mobile_state [, target_state | ||
| + | [, quiet [, max_skip [, transform [, reset ]]]]]]]]]]]]] | ||
| + | |||
| + | == Arguments == | ||
| + | |||
| + | * '''mobile''' = string: atom selection of mobile object | ||
| + | * '''target''' = string: atom selection of target object | ||
| + | * '''cutoff''' = float: outlier rejection cutoff in RMS {default: 2.0} | ||
| + | * '''cycles''' = int: maximum number of outlier rejection cycles {default: 5} | ||
| + | * '''gap, extend, max_gap''': sequence alignment parameters | ||
| + | * '''object''' = string: name of alignment object to create {default: (no alignment object)} | ||
| + | * '''matrix''' = string: file name of substitution matrix for sequence alignment {default: BLOSUM62} | ||
| + | * '''mobile_state''' = int: object state of mobile selection {default: 0 = all states} | ||
| + | * '''target_state''' = int: object state of target selection {default: 0 = all states} | ||
| + | * '''quiet''' = 0/1: suppress output {default: 0 in command mode, 1 in API} | ||
| + | * '''max_skip''' = ? | ||
| + | * '''transform''' = 0/1: do superposition {default: 1} | ||
| + | * '''reset''' = ? | ||
| + | |||
| + | == Alignment Objects == | ||
| + | |||
| + | An alignment object can be created with the '''object='''''somename'' argument. An alignment object provides: | ||
| + | |||
| + | * aligned [[seq_view|sequence viewer]] | ||
| + | * graphical representation of aligned atom pairs as lines in the 3D viewer | ||
| + | * can be [[save|saved]] to a clustalw sequence alignment file | ||
| + | |||
| + | == RMSD == | ||
| + | |||
| + | The RMSD of the aligned atoms (after outlier rejection!) is reported in the text output. The '''all-atom RMSD''' can be obtained by setting '''cycles=0''' and thus not doing any outlier rejection. The RMSD can also be captured with a python script, see the [[#PyMOL API|API paragraph]] below. Note that the output prints "RMS" but it is in fact "RMSD" and the units are Angstroms. | ||
| + | |||
| + | == Examples == | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | fetch 1oky 1t46, async=0 | ||
| + | |||
| + | # 1) default with outlier rejection | ||
| + | align 1oky, 1t46 | ||
| + | |||
| + | # 2) with alignment object, save to clustalw file | ||
| + | align 1oky, 1t46, object=alnobj | ||
| + | save alignment.aln, alnobj | ||
| + | |||
| + | # 3) all-atom RMSD (no outlier rejection) and without superposition | ||
| + | align 1oky, 1t46, cycles=0, transform=0 | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | == PyMOL API == | ||
<source lang="python"> | <source lang="python"> | ||
| − | + | cmd.align( string mobile, string target, float cutoff=2.0, | |
| − | + | int cycles=5, float gap=-10.0, float extend=-0.5, | |
| + | int max_gap=50, string object=None, string matrix='BLOSUM62', | ||
| + | int mobile_state=0, int target_state=0, int quiet=1, | ||
| + | int max_skip=0, int transform=1, int reset=0 ) | ||
</source> | </source> | ||
| − | + | This returns a list with 7 items: | |
| − | + | ||
| − | + | # RMSD after refinement | |
| − | + | # Number of aligned atoms after refinement | |
| − | + | # Number of refinement cycles | |
| − | + | # RMSD before refinement | |
| − | + | # Number of aligned atoms before refinement | |
| + | # Raw alignment score | ||
| + | # Number of residues aligned | ||
| + | |||
| + | == Notes == | ||
| − | + | * The molecules you want to align need to be in '''two different objects'''. Else, PyMOL will answer with: ''ExecutiveAlign: invalid selections for alignment.'' You can skirt this problem by making a temporary object and aligning your original to the copy. | |
| − | + | * By defaults, '''all states''' (like in NMR structures or trajectories) are considered, this might yield a bad or suboptimal alignment for a single state. Use the '''mobile_state''' and '''target_state''' argument to be explicit in such cases. | |
| − | |||
| − | === | + | == See Also == |
| − | |||
| − | |||
| − | |||
| − | + | * [[super]], [[cealign]], [[fit]], [[pair_fit]] | |
| − | + | * [[rms]], [[rms_cur]], | |
| + | * [[intra_fit]], [[intra_rms]], [[intra_rms_cur]] | ||
| + | * [[extra_fit]] | ||
| + | * [http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/ align_all.py and super_all.py] | ||
| + | * [[tmalign]] | ||
| + | * [[Color_by_conservation]] | ||
| + | * [[Get_raw_alignment]] | ||
| + | * [[mcsalign]] (psico) | ||
| − | [[Category:Commands| | + | [[Category:Commands|Align]] |
| + | [[Category:Structure_Alignment|Align]] | ||
Latest revision as of 04:50, 11 December 2017
align performs a sequence alignment followed by a structural superposition, and then carries out zero or more cycles of refinement in order to reject structural outliers found during the fit. align does a good job on proteins with decent sequence similarity (identity >30%). For comparing proteins with lower sequence identity, the super and cealign commands perform better.
Usage
align mobile, target [, cutoff [, cycles
[, gap [, extend [, max_gap [, object
[, matrix [, mobile_state [, target_state
[, quiet [, max_skip [, transform [, reset ]]]]]]]]]]]]]
Arguments
- mobile = string: atom selection of mobile object
- target = string: atom selection of target object
- cutoff = float: outlier rejection cutoff in RMS {default: 2.0}
- cycles = int: maximum number of outlier rejection cycles {default: 5}
- gap, extend, max_gap: sequence alignment parameters
- object = string: name of alignment object to create {default: (no alignment object)}
- matrix = string: file name of substitution matrix for sequence alignment {default: BLOSUM62}
- mobile_state = int: object state of mobile selection {default: 0 = all states}
- target_state = int: object state of target selection {default: 0 = all states}
- quiet = 0/1: suppress output {default: 0 in command mode, 1 in API}
- max_skip = ?
- transform = 0/1: do superposition {default: 1}
- reset = ?
Alignment Objects
An alignment object can be created with the object=somename argument. An alignment object provides:
- aligned sequence viewer
- graphical representation of aligned atom pairs as lines in the 3D viewer
- can be saved to a clustalw sequence alignment file
RMSD
The RMSD of the aligned atoms (after outlier rejection!) is reported in the text output. The all-atom RMSD can be obtained by setting cycles=0 and thus not doing any outlier rejection. The RMSD can also be captured with a python script, see the API paragraph below. Note that the output prints "RMS" but it is in fact "RMSD" and the units are Angstroms.
Examples
fetch 1oky 1t46, async=0
# 1) default with outlier rejection
align 1oky, 1t46
# 2) with alignment object, save to clustalw file
align 1oky, 1t46, object=alnobj
save alignment.aln, alnobj
# 3) all-atom RMSD (no outlier rejection) and without superposition
align 1oky, 1t46, cycles=0, transform=0
PyMOL API
cmd.align( string mobile, string target, float cutoff=2.0,
int cycles=5, float gap=-10.0, float extend=-0.5,
int max_gap=50, string object=None, string matrix='BLOSUM62',
int mobile_state=0, int target_state=0, int quiet=1,
int max_skip=0, int transform=1, int reset=0 )
This returns a list with 7 items:
- RMSD after refinement
- Number of aligned atoms after refinement
- Number of refinement cycles
- RMSD before refinement
- Number of aligned atoms before refinement
- Raw alignment score
- Number of residues aligned
Notes
- The molecules you want to align need to be in two different objects. Else, PyMOL will answer with: ExecutiveAlign: invalid selections for alignment. You can skirt this problem by making a temporary object and aligning your original to the copy.
- By defaults, all states (like in NMR structures or trajectories) are considered, this might yield a bad or suboptimal alignment for a single state. Use the mobile_state and target_state argument to be explicit in such cases.