This is a read-only mirror of pymolwiki.org
Difference between revisions of "Radius of gyration"
Jump to navigation
Jump to search
m |
m (1 revision) |
||
| (5 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Infobox psico | ||
| + | |module = psico.querying#gyradius | ||
| + | }} | ||
| + | |||
This script does calcualate the [http://en.wikipedia.org/wiki/Radius_of_gyration radius of gyration] of a molecule. | This script does calcualate the [http://en.wikipedia.org/wiki/Radius_of_gyration radius of gyration] of a molecule. | ||
Thanks to Tsjerk Wassenaar for posting this script on the [http://sourceforge.net/mailarchive/message.php?msg_id=27288491 pymol-users mailing list]! | Thanks to Tsjerk Wassenaar for posting this script on the [http://sourceforge.net/mailarchive/message.php?msg_id=27288491 pymol-users mailing list]! | ||
| + | |||
| + | ''The function was added to [[psico]] as "gyradius".'' | ||
<source lang="python"> | <source lang="python"> | ||
from pymol import cmd | from pymol import cmd | ||
| − | |||
import math | import math | ||
| Line 18: | Line 23: | ||
rgyrate [ selection ] | rgyrate [ selection ] | ||
''' | ''' | ||
| + | try: | ||
| + | from itertools import izip | ||
| + | except ImportError: | ||
| + | izip = zip | ||
quiet = int(quiet) | quiet = int(quiet) | ||
model = cmd.get_model(selection).atom | model = cmd.get_model(selection).atom | ||
| Line 28: | Line 37: | ||
rg = math.sqrt(rr/tmass - mm) | rg = math.sqrt(rr/tmass - mm) | ||
if not quiet: | if not quiet: | ||
| − | print "Radius of gyration: %.2f" % (rg) | + | print("Radius of gyration: %.2f" % (rg)) |
return rg | return rg | ||
| Line 52: | Line 61: | ||
cmd.show('cartoon', 'chain A') | cmd.show('cartoon', 'chain A') | ||
| − | + | r = radiusOfGyration.rgyrate('chain A and polymer') | |
| − | centerOfMass.com('chain A and polymer', object=' | + | centerOfMass.com('chain A and polymer', object='com', vdw=r) |
| − | |||
util.cbc() | util.cbc() | ||
| Line 61: | Line 69: | ||
=== See Also === | === See Also === | ||
| − | [[Center Of Mass]] | + | * [[centerofmass]] |
| + | * [[Center Of Mass]] | ||
| + | * [[COM]] | ||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:Math_Scripts]] | [[Category:Math_Scripts]] | ||
Latest revision as of 02:27, 28 May 2019
|
Included in psico | |
| Module | psico.querying#gyradius |
|---|---|
This script does calcualate the radius of gyration of a molecule.
Thanks to Tsjerk Wassenaar for posting this script on the pymol-users mailing list!
The function was added to psico as "gyradius".
from pymol import cmd
import math
def rgyrate(selection='(all)', quiet=1):
'''
DESCRIPTION
Radius of gyration
USAGE
rgyrate [ selection ]
'''
try:
from itertools import izip
except ImportError:
izip = zip
quiet = int(quiet)
model = cmd.get_model(selection).atom
x = [i.coord for i in model]
mass = [i.get_mass() for i in model]
xm = [(m*i,m*j,m*k) for (i,j,k),m in izip(x,mass)]
tmass = sum(mass)
rr = sum(mi*i+mj*j+mk*k for (i,j,k),(mi,mj,mk) in izip(x,xm))
mm = sum((sum(i)/tmass)**2 for i in izip(*xm))
rg = math.sqrt(rr/tmass - mm)
if not quiet:
print("Radius of gyration: %.2f" % (rg))
return rg
cmd.extend("rgyrate", rgyrate)
# vi:expandtab
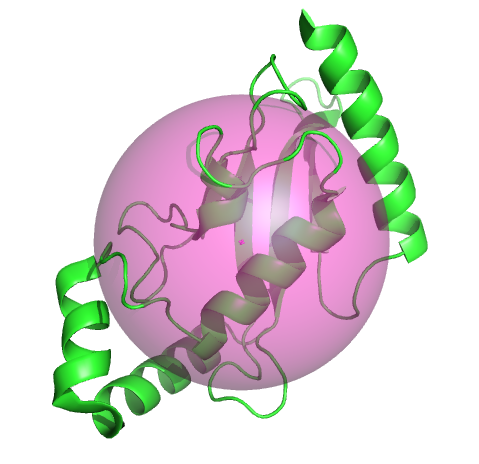
Example
This example uses the radius of gyration and the center of mass to display a semitransparent sphere.
from pymol import cmd, stored, util
import centerOfMass, radiusOfGyration
cmd.set('sphere_transparency', 0.5)
cmd.fetch('2xwu')
cmd.hide('everything')
cmd.show('cartoon', 'chain A')
r = radiusOfGyration.rgyrate('chain A and polymer')
centerOfMass.com('chain A and polymer', object='com', vdw=r)
util.cbc()