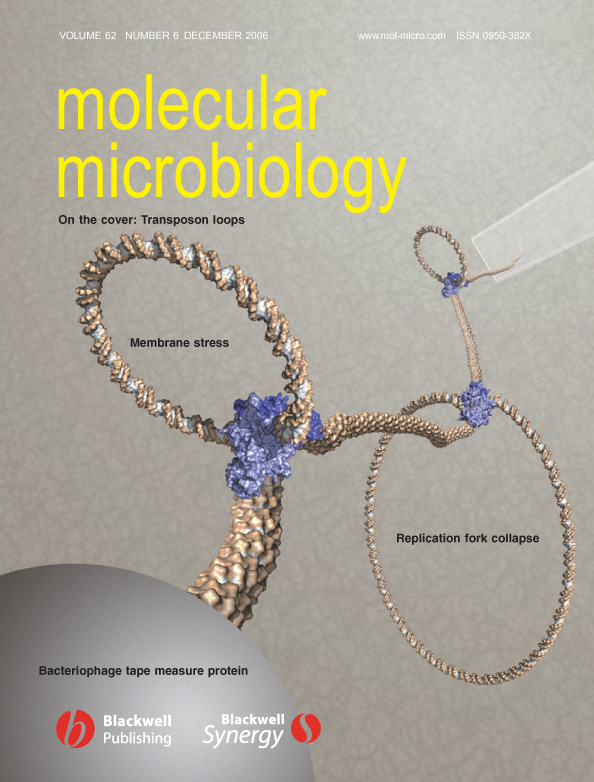
| Official Release
|
PyMOL v2.0 has been released on September 20, 2017.
|
| Plugin Update
|
MOLE 2.5 is an updated version of channel analysis software in PyMOL
|
| Official Release
|
PyMOL v1.8.6 has been released on March 9, 2017.
|
| Official Release
|
PyMOL v1.8.4 has been released on October 4, 2016.
|
| New Script
|
dssr_block is a wrapper for DSSR (3dna) and creates block-shaped nucleic acid cartoons
|
| New Plugin
|
LiSiCA is a new plugin for 2D and 3D ligand based virtual screening using a fast maximum clique algorithm.
|
| Official Release
|
PyMOL v1.8.0 has been released on Nov 18, 2015.
|
| PyMOL Open-Source Fellowship
|
Schrödinger is now accepting applications for the PyMOL Open-Source Fellowship program! Details on http://pymol.org/fellowship
|
| Official Release
|
PyMOL, AxPyMOL, and JyMOL v1.7.6 have all been released on May 4, 2015.
|
| New Plugin
|
PyANM is a new plugin for easier Anisotropic Network Model (ANM) building and visualising in PyMOL.
|
| New Plugin
|
Bondpack is a collection of PyMOL plugins for easy visualization of atomic bonds.
|
| New Plugin
|
MOLE 2.0 is a new plugin for rapid analysis of biomacromolecular channels in PyMOL.
|
| 3D using Geforce
|
PyMOL can now be visualized in 3D using Nvidia GeForce video cards (series 400+) with 120Hz monitors and Nvidia 3D Vision, this was previously only possible with Quadro video cards.
|
| Older News
|
See Older News.
|