This is a read-only mirror of pymolwiki.org
Lisica
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/lisica.py |
| Author(s) | Janez Konc |
| License | |
| This code has been put under version control in the project Pymol-script-repo | |
Important
Currently, the most recent version can be obtained at the Insilab web page.
Description
LiSiCA is a software for 2D and 3D ligand based virtual screening. It uses a fast maximum clique algorithm to find two- and three- dimensional similarities between reference compound and a database of target compounds in Mol2 format. The similarities are expressed using Tanimoto coefficients and the target compounds are ranked accordingly. LiSiCA is developed and maintained at National Institute of Chemistry, Slovenia.
LiSiCA Plugin is free for Academic (NON-COMMERCIAL) use. However, for COMMERCIAL use the potential users have to write to Janez Konc.
LiSiCA Plugin Features
- Graphical User Interface which facilitate the choice of parameters for LiSiCA.
- Results displayed according to the ranking in similarities as measured by Tanimoto coefficients.
- Structural similarity between molecules visualized using PyMOL viewer.
Requirements
The plugin should work on Windows and all variety of Unix like systems. Most of the tests were performed on Windows 7, Ubuntu 14.04 and Linux Mint. To work properly, the plugin requires Python with Tkinter and PyMOL. The plugin was mainly tested with PyMOL 1.7.x.
Installation
This plugin is ready "out-of-box" in most Linux and Windows operating systems and is available on GitHub in the Pymol-script-repo repository.
The lisica.py script initializes the installation of the plugin. The script downloads and installs all the required files. On successful installation, a directory named .lisicagui is downloaded and saved in the home directory. This folder contains the executables, log files, icon files, python modules etc.
Once the files are properly installed, LiSiCA plugin version will be ready to use.
Important note: If installation fails with "pmg_tk" error or similar, one or both of the tkinter and ttk libraries are not installed. To install them open terminal and type as root "yum install tkinter python-pip" and "pip install pyttk" (in CentOS) or "apt-get install tkinter python-pip" and then "pip install pyttk" (in Ubuntu).
Important note: For complete installation of the plugin, a stable INTERNET connection is required.
Usage
The plugin window has four tabs.
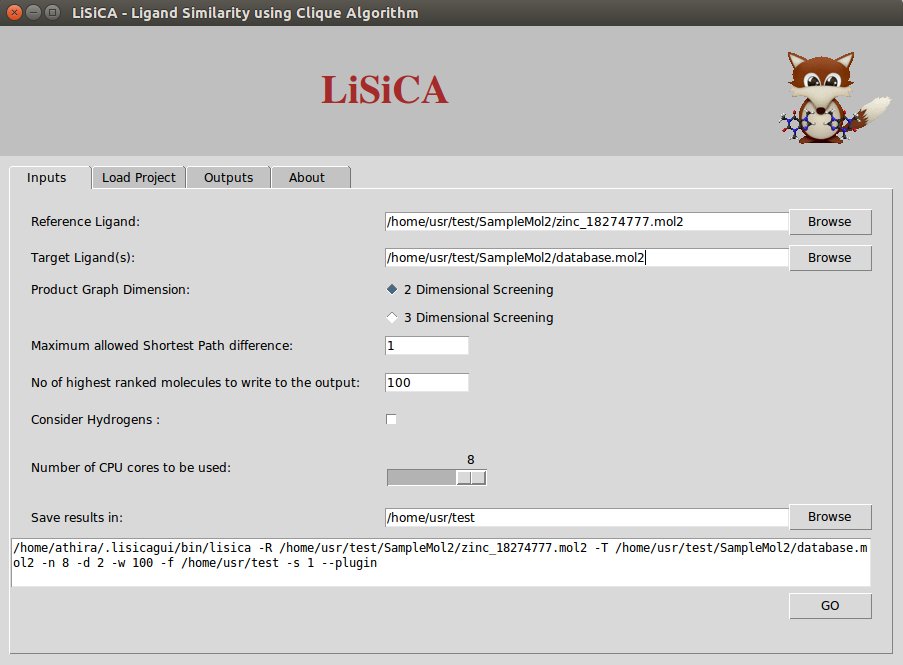
Inputs Tab
Both the reference and the target compound files need to be in the Tripos mol2 format. The reference file should contain only one (reference) compound; the target file may contain many compounds. If the reference file contains more than one compound, the first molecule is used as a reference. The molecules in the target file having the same name are considered as different conformers of the same molecule. By default only the best-scoring (by Tanimoto coefficient) conformer will be shown in the final output for the 3D screening option.
In the mol2 input files, the name of the molecule (ZINC ID) must be specified under the line for @<TRIPOS>MOLECULE tag. For example,
@<TRIPOS>MOLECULE ZINC73655097 46 48 0 0 0 SMALL USER_CHARGES @<TRIPOS>ATOM ...
LiSiCA checks similarities based on the mol2 atom types. Hence SYBYL atom types have to be specified in the mol2 files. For example, under the @<TRIPOS>ATOM tag, each atom specification should include the SYBYL atom type, as in:
1 C1 -0.0647 1.4496 -0.0592 C.3 1 <0> -0.167
In the above line from a mol2 file, in bold, in column 5, the SYBYL atom type is specified.
In 2D screening, the option Maximum Allowed Shortest path Difference corresponds to the maximum allowed difference in shortest-path length between
atoms of the two compared product graph vertices. Lesser values correspond to a more rigorous screening. By default this value is unit bond.
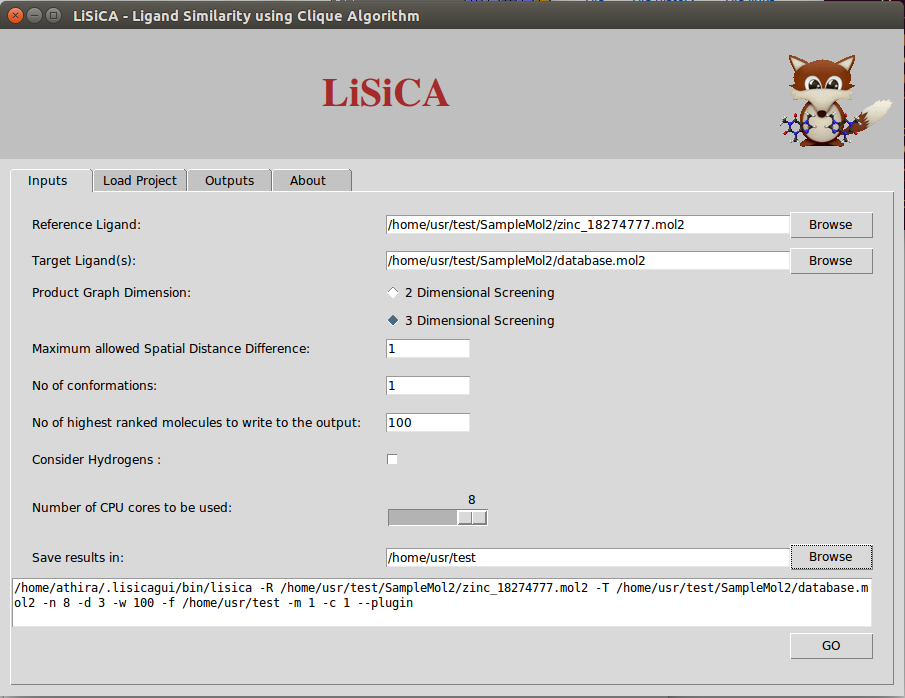
In 3D screening, the option Maximum Allowed Spatial Distance Difference corresponds to the maximum allowed difference in distances between atoms of the two compared product graph vertices. Lesser values correspond to a more rigorous screening. By default this value is 1 Å. The Number of Conformations option corresponds to the maximum number of outputted files of one molecule in different conformations and is to be used only for 3D screening.
According to the value of Number of highest ranked molecules to be written to the output (say W), LiSiCA will create mol2 files of that many (W) highest scoring
target molecules with a comment section at the end of the file where the matching atom pairs are displayed. This value is by default 100. The resulting mol2 files
are written into a time-stamp directory in the folder specified in Save results in:. By default this folder is the user's home directory. Also, a text file named
lisica_results.txt with the target molecules with (in the descending order of) Tanimoto coefficients is written to the time-stamp folder.
The Number of CPU cores to be used allows selection of CPU threads used for LiSiCA. By default, it tries to detect the number of CPUs available. The Consider Hydrogen options lets the user to choose if the hydrogen atoms are to be considered for the calculation of the similarity using the maximum clique algorithm. By default, hydrogen atoms are not considered in finding the largest substructure common to the reference and target molecules, so as to obtain faster results.
Load Project Tab
The plugin also has a feature to load saved results. On the Load Project tab, the user can choose the directory with the saved results (mol2 files of each target and the reference) and the lisica_results.txt file. When the load button is clicked, the results will be loaded onto the output tab and the PyMOL Viewer window.
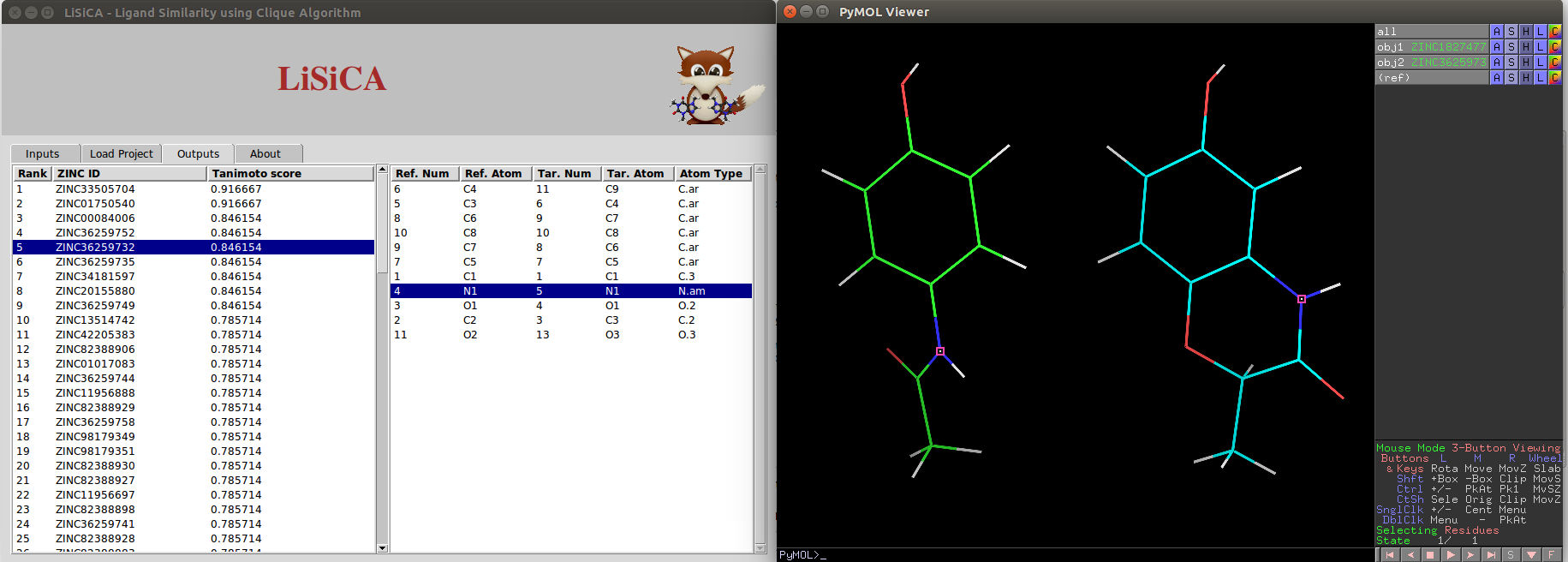
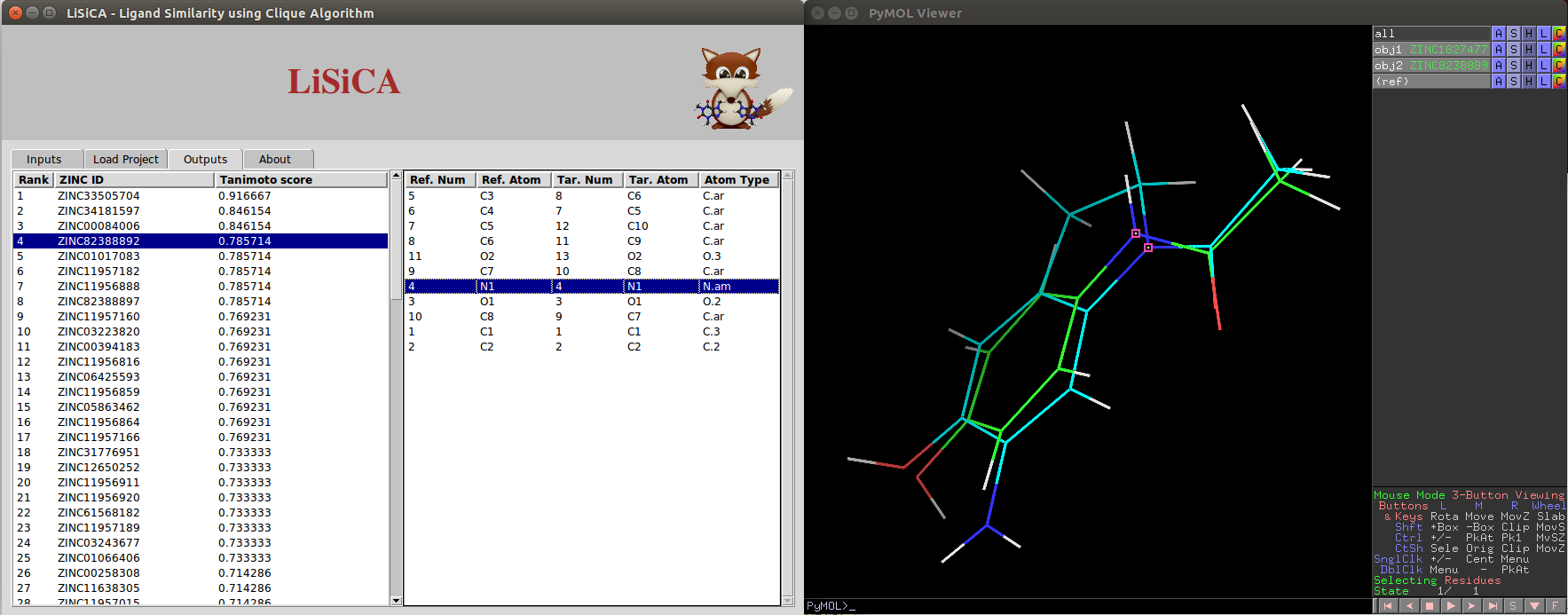
Output Tab
In the output tab, there are two listboxes:
- One contains ZINC ID and Tanimoto Coefficients of target molecules in the decreasing order of the Tanimoto coefficient values.
Any single target molecule can be selected on this listbox using a mouse click or using up/down arrow keys. The selected target molecule is displayed with the reference molecule on the PyMOL viewer window.
- Depending on the target molecule chosen on the first listbox, the corresponding atoms from reference and the target molecules are displayed on the other listbox.
Any single pair of corresponding atoms can be selected on this listbox using a mouse click or using up/down arrow keys. The selected pair is highlighted on the PyMOL viewer window.
For 2D Screening, the two molecules (the reference and the selected target) are visualized side by side on the PyMOL viewer screen.
For 3D Screening, the 3D structures of the two molecules (the reference and the selected target) are superimposed on one another to visualize the similarity on the PyMOL viewer screen.
About Tab
The users can get information on new updates if available on the About tab.
Reference
If you are using LiSiCA in your work, please cite:
S. Lesnik, T. Stular, B. Brus, D. Knez, S. Gobec, D. Janezic, J. Konc, LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors, J. Chem. Inf. Model., 2015, 55, 1521–1528.
Licensing
LiSiCA software is copyrighted by :
- Janez Konc
- National Institute of Chemistry
- Laboratory for Molecular Modeling
- Hajdrihova 19
- SI-1000 Ljubljana
- Slovenia.
The terms stated in the following agreement apply to all files associated with the software unless explicitly disclaimed in individual files.
LiSiCA SOFTWARE LICENSE AGREEMENT
1. Grant Of Limited License; Software Use Restrictions. The programs received by you will be used only for NON COMMERCIAL purposes.
This license is issued to you as an individual.
For COMMERCIAL use of the software, please contact Janez Konc for details about commercial usage license agreements.
For any question regarding license agreements, please contact:
Janez Konc
National Institute of Chemistry
Laboratory for Molecular Modeling
Hajdrihova 19
SI-1000 Ljubljana
Slovenia.
2. COMMERCIAL USAGE is defined as revenues generating activities. These
include using this software for consulting activities and selling
applications built on top of, or using this software. Scientific
research in an academic environment and teaching are considered
NON COMMERCIAL.
3. Copying Restrictions. You will not sell or otherwise distribute commercially
these programs or derivatives to any other party, whether with or without
consideration.
4. Ownership of Software. You will not obtain, and will not attempt to
obtain copyright coverage thereon without the express purpose written
consent of Janez Konc.
5. IN NO EVENT SHALL THE AUTHORS OR DISTRIBUTORS BE LIABLE TO ANY PARTY
FOR DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES
ARISING OUT OF THE USE OF THIS SOFTWARE, ITS DOCUMENTATION, OR ANY
DERIVATIVES THEREOF, EVEN IF THE AUTHORS HAVE BEEN ADVISED OF THE
POSSIBILITY OF SUCH DAMAGE.
6. THE AUTHORS AND DISTRIBUTORS SPECIFICALLY DISCLAIM ANY WARRANTIES,
INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE, AND NON-INFRINGEMENT. THIS SOFTWARE
IS PROVIDED ON AN "AS IS" BASIS, AND THE AUTHORS AND DISTRIBUTORS HAVE
NO OBLIGATION TO PROVIDE MAINTENANCE, SUPPORT, UPDATES, ENHANCEMENTS, OR
MODIFICATIONS.