This is a read-only mirror of pymolwiki.org
Difference between revisions of "Selection Macros"
m (1 revision) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
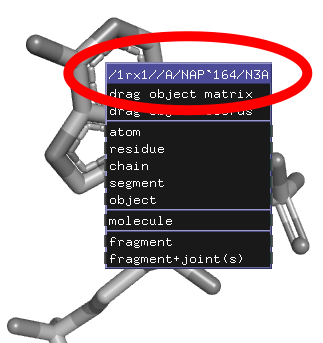
| − | + | [[Image:right-click-macro.png|right|frame|The atom-right-click context menu title shows a selection macro]] | |
| − | + | ||
| − | Macros | + | [[Selection Macros]] allow to represent a long [[Selection Algebra|atom selection]] phrase such as |
| − | + | PyMOL> select pept and segi lig and chain B and resi 142 and name CA | |
in a more compact form: | in a more compact form: | ||
| − | + | PyMOL> select /pept/lig/B/142/CA | |
| + | |||
| + | == Syntax and Semantics == | ||
An atom selection macro uses slashes to define fields corresponding to identifiers. The macro is used to select atoms using the boolean "and," that is, the selected atoms must have all the matching identifiers: | An atom selection macro uses slashes to define fields corresponding to identifiers. The macro is used to select atoms using the boolean "and," that is, the selected atoms must have all the matching identifiers: | ||
| − | < | + | <source lang="python"> |
| + | /object-name/segi-identifier/chain-identifier/resi-identifier/name-identifier | ||
| + | </source> | ||
| + | |||
| + | * must contain '''at least one slash''' | ||
| + | * no spaces allowed | ||
| + | * '''empty''' fields are interpreted as '''wildcards''' | ||
| + | * starting with a slash: | ||
| + | ** '''yes''': fields from the '''right''' can be omitted | ||
| + | ** '''no''': fields from the '''left''' can be omitted | ||
| − | + | == Details == | |
Macros come in two flavors: those that begin with a slash and those that don't. The presence or absence of a slash at the beginning of the macro determines how it is interpreted. If the macro begins with a slash, PyMOL expects to find the fields starting from the top of the hierarchy: the first field to the right of the slash is interpreted as an object-name; the second field as an identifier to segi; the third as an identifier to chain, and so on. It may take any of the following forms: | Macros come in two flavors: those that begin with a slash and those that don't. The presence or absence of a slash at the beginning of the macro determines how it is interpreted. If the macro begins with a slash, PyMOL expects to find the fields starting from the top of the hierarchy: the first field to the right of the slash is interpreted as an object-name; the second field as an identifier to segi; the third as an identifier to chain, and so on. It may take any of the following forms: | ||
| Line 29: | Line 40: | ||
PyMOL> zoom /pept | PyMOL> zoom /pept | ||
PyMOL> show spheres, /pept/lig/ | PyMOL> show spheres, /pept/lig/ | ||
| − | PyMOL> show cartoon, /pept/lig/ | + | PyMOL> show cartoon, /pept/lig/A |
| − | PyMOL> color pink, /pept/lig/ | + | PyMOL> color pink, /pept/lig/A/10 |
| − | PyMOL> color yellow, /pept/lig/ | + | PyMOL> color yellow, /pept/lig/A/10/CA |
</pre> | </pre> | ||
| Line 44: | Line 55: | ||
EXAMPLES | EXAMPLES | ||
| − | PyMOL> zoom 10/ | + | PyMOL> zoom 10/CB |
| − | PyMOL> show spheres, | + | PyMOL> show spheres, A/10-12/CB |
| − | PyMOL> show cartoon, lig/ | + | PyMOL> show cartoon, lig/B/6+8/C+O |
| − | PyMOL> color pink, pept/enz/ | + | PyMOL> color pink, pept/enz/C/3/N |
</pre> | </pre> | ||
| − | |||
===Omitting Fields in a Macro=== | ===Omitting Fields in a Macro=== | ||
| Line 64: | Line 74: | ||
PyMOL> zoom 142/ # Residue 142 fills the viewer. | PyMOL> zoom 142/ # Residue 142 fills the viewer. | ||
| − | PyMOL> show spheres, 156/ | + | PyMOL> show spheres, 156/CA # The alpha carbon of residue 156 |
# is shown as a sphere | # is shown as a sphere | ||
| − | PyMOL> show cartoon, | + | PyMOL> show cartoon, A// # Chain "A" is shown as a cartoon. |
| − | PyMOL> color pink, pept// | + | PyMOL> color pink, /pept//B # Chain "B" in object "pept" |
# is colored pink. | # is colored pink. | ||
</pre> | </pre> | ||
| + | == See Also == | ||
| − | + | * [[Selection Algebra]] | |
| − | Selection | ||
| − | |||
[[Category:Selector Quick Reference]] | [[Category:Selector Quick Reference]] | ||
| − | + | [[Category:Selecting|Selection Macros]] | |
| − | [[Category: | + | [[Category:Coloring]] |
Latest revision as of 04:50, 11 December 2017
Selection Macros allow to represent a long atom selection phrase such as
PyMOL> select pept and segi lig and chain B and resi 142 and name CA
in a more compact form:
PyMOL> select /pept/lig/B/142/CA
Syntax and Semantics
An atom selection macro uses slashes to define fields corresponding to identifiers. The macro is used to select atoms using the boolean "and," that is, the selected atoms must have all the matching identifiers:
/object-name/segi-identifier/chain-identifier/resi-identifier/name-identifier
- must contain at least one slash
- no spaces allowed
- empty fields are interpreted as wildcards
- starting with a slash:
- yes: fields from the right can be omitted
- no: fields from the left can be omitted
Details
Macros come in two flavors: those that begin with a slash and those that don't. The presence or absence of a slash at the beginning of the macro determines how it is interpreted. If the macro begins with a slash, PyMOL expects to find the fields starting from the top of the hierarchy: the first field to the right of the slash is interpreted as an object-name; the second field as an identifier to segi; the third as an identifier to chain, and so on. It may take any of the following forms:
Macros Beginning With a Slash
/object-name/segi-identifier/chain-identifier/resi-identifier/name-identifier /object-name/segi-identifier/chain-identifier/resi-identifier /object-name/segi-identifier/chain-identifier /object-name/segi-identifier /object-name EXAMPLES PyMOL> zoom /pept PyMOL> show spheres, /pept/lig/ PyMOL> show cartoon, /pept/lig/A PyMOL> color pink, /pept/lig/A/10 PyMOL> color yellow, /pept/lig/A/10/CA
Macros Not Beginning With a Slash
If the macro does not begin with a slash, it is interpreted differently. In this case, PyMOL expects to find the fields ending with the bottom of the hierarchy. Macros that don't start with a slash may take the following forms:
resi-identifier/name-identifier
chain-identifier/resi-identifier/name-identifier
segi-identifier/chain-identifier/resi-identifier/name-identifier
object-name/segi-identifier/chain-identifier/resi-identifier/name-identifier
EXAMPLES
PyMOL> zoom 10/CB
PyMOL> show spheres, A/10-12/CB
PyMOL> show cartoon, lig/B/6+8/C+O
PyMOL> color pink, pept/enz/C/3/N
Omitting Fields in a Macro
You can also omit fields between slashes. Omitted fields will be interpreted as wildcards, as in the following forms:
resi-identifier/
resi-identifier/name-identifier
chain-identifier//
object-name//chain-identifier
EXAMPLES
PyMOL> zoom 142/ # Residue 142 fills the viewer.
PyMOL> show spheres, 156/CA # The alpha carbon of residue 156
# is shown as a sphere
PyMOL> show cartoon, A// # Chain "A" is shown as a cartoon.
PyMOL> color pink, /pept//B # Chain "B" in object "pept"
# is colored pink.