|
|
| (41 intermediate revisions by one other user not shown) |
| Line 1: |
Line 1: |
| − | == Author == | + | {{Infobox script-repo |
| − | This pymol script is made by Troels Emtekær Linnet
| + | |type = script |
| | + | |filename = rotkit.py |
| | + | |author = [[User:Tlinnet|Troels E. Linnet]] |
| | + | |license = BSD |
| | + | }} |
| | | | |
| | == Introduction == | | == Introduction == |
| | + | This script-kit is a collection of small script to be able to precisely to put a molecule (like a dye) where you want in relation to a protein. |
| | | | |
| | + | You can also create rotational states of a domain or simulate a dye freedom. |
| | | | |
| − | == Example of use ==
| + | It simply makes the [[Object_Matrix | PyMOL TTT matrixes]], in a easy and user friendly way. The calls to the functions available in PyMOL, takes care of all the |
| | + | conversion of input and such. |
| | | | |
| − | === The dye .pdb file ===
| + | If you are interested in this, you might also want to check out the PyMOL [[Chempy]] module that is included in PyMOL. It provides handy vector and matrix functions. |
| − | <source lang="python">
| |
| − | COMPND Atto590
| |
| − | AUTHOR GENERATED BY OPEN BABEL 2.2.3 an in Avogadro
| |
| − | HETATM 1 C LIG 1 -4.057 2.454 -1.838 1.00 0.00 C
| |
| − | HETATM 2 N LIG 1 -2.619 2.797 -1.904 1.00 0.00 N
| |
| − | HETATM 3 C LIG 1 -4.780 3.335 -0.830 1.00 0.00 C
| |
| − | HETATM 4 C LIG 1 -4.192 1.001 -1.344 1.00 0.00 C
| |
| − | HETATM 5 C LIG 1 -4.793 2.597 -3.193 1.00 0.00 C
| |
| − | HETATM 6 C LIG 1 -1.942 3.160 -0.686 1.00 0.00 C
| |
| − | HETATM 7 C LIG 1 -4.149 3.889 0.224 1.00 0.00 C
| |
| − | HETATM 8 C LIG 1 -2.678 3.711 0.383 1.00 0.00 C
| |
| − | HETATM 9 C LIG 1 -2.011 4.104 1.567 1.00 0.00 C
| |
| − | HETATM 10 C LIG 1 -0.555 2.983 -0.540 1.00 0.00 C
| |
| − | HETATM 11 C LIG 1 -0.625 3.896 1.713 1.00 0.00 C
| |
| − | HETATM 12 C LIG 1 0.087 3.328 0.645 1.00 0.00 C
| |
| − | HETATM 13 O LIG 1 1.412 3.104 0.756 1.00 0.00 O
| |
| − | HETATM 14 C LIG 1 0.146 4.247 2.961 1.00 0.00 C
| |
| − | HETATM 15 C LIG 1 1.477 3.899 3.020 1.00 0.00 C
| |
| − | HETATM 16 C LIG 1 2.103 3.331 1.896 1.00 0.00 C
| |
| − | HETATM 17 C LIG 1 3.458 3.002 1.923 1.00 0.00 C
| |
| − | HETATM 18 C LIG 1 2.254 4.083 4.182 1.00 0.00 C
| |
| − | HETATM 19 C LIG 1 4.257 3.179 3.087 1.00 0.00 C
| |
| − | HETATM 20 C LIG 1 3.617 3.727 4.231 1.00 0.00 C
| |
| − | HETATM 21 N LIG 1 5.544 2.857 3.105 1.00 0.00 N
| |
| − | HETATM 22 C LIG 1 4.377 3.930 5.483 1.00 0.00 C
| |
| − | HETATM 23 C LIG 1 6.402 3.070 4.308 1.00 0.00 C
| |
| − | HETATM 24 C LIG 1 5.668 3.624 5.509 1.00 0.00 C
| |
| − | HETATM 25 C LIG 1 7.010 1.725 4.768 1.00 0.00 C
| |
| − | HETATM 26 C LIG 1 7.526 4.086 3.998 1.00 0.00 C
| |
| − | HETATM 27 H LIG 1 -5.861 3.428 -0.908 1.00 0.00 H
| |
| − | HETATM 28 H LIG 1 -2.574 4.547 2.368 1.00 0.00 H
| |
| − | HETATM 29 H LIG 1 0.038 2.543 -1.329 1.00 0.00 H
| |
| − | HETATM 30 C LIG 1 -0.510 4.983 4.109 1.00 0.00 C
| |
| − | HETATM 31 H LIG 1 1.794 4.486 5.067 1.00 0.00 H
| |
| − | HETATM 32 H LIG 1 3.861 2.612 1.004 1.00 0.00 H
| |
| − | HETATM 33 H LIG 1 6.234 3.777 6.426 1.00 0.00 H
| |
| − | HETATM 34 C LIG 1 3.721 4.478 6.733 1.00 0.00 C
| |
| − | HETATM 35 H LIG 1 6.199 1.004 5.010 1.00 0.00 H
| |
| − | HETATM 36 H LIG 1 7.638 1.861 5.675 1.00 0.00 H
| |
| − | HETATM 37 H LIG 1 7.662 1.271 3.996 1.00 0.00 H
| |
| − | HETATM 38 H LIG 1 7.085 5.060 3.696 1.00 0.00 H
| |
| − | HETATM 39 H LIG 1 8.209 3.753 3.194 1.00 0.00 H
| |
| − | HETATM 40 H LIG 1 8.161 4.256 4.897 1.00 0.00 H
| |
| − | HETATM 41 C LIG 1 6.193 2.277 1.907 1.00 0.00 C
| |
| − | HETATM 42 H LIG 1 6.034 2.929 1.021 1.00 0.00 H
| |
| − | HETATM 43 C LIG 1 5.765 0.821 1.653 1.00 0.00 C
| |
| − | HETATM 44 H LIG 1 7.291 2.219 2.020 1.00 0.00 H
| |
| − | HETATM 45 H LIG 1 4.681 0.690 1.481 1.00 0.00 H
| |
| − | HETATM 46 H LIG 1 6.040 0.186 2.519 1.00 0.00 H
| |
| − | HETATM 47 H LIG 1 6.299 0.437 0.758 1.00 0.00 H
| |
| − | HETATM 48 C LIG 1 -1.890 2.833 -3.182 1.00 0.00 C
| |
| − | HETATM 49 H LIG 1 -2.440 3.435 -3.934 1.00 0.00 H
| |
| − | HETATM 50 C LIG 1 -1.575 1.431 -3.708 1.00 0.00 C
| |
| − | HETATM 51 H LIG 1 -0.922 3.362 -3.078 1.00 0.00 H
| |
| − | HETATM 52 H LIG 1 -2.484 0.841 -3.923 1.00 0.00 H
| |
| − | HETATM 53 H LIG 1 -0.963 0.879 -2.964 1.00 0.00 H
| |
| − | HETATM 54 H LIG 1 -0.995 1.515 -4.651 1.00 0.00 H
| |
| − | HETATM 55 H LIG 1 -4.746 3.648 -3.551 1.00 0.00 H
| |
| − | HETATM 56 H LIG 1 -5.866 2.324 -3.085 1.00 0.00 H
| |
| − | HETATM 57 H LIG 1 -4.381 1.935 -3.977 1.00 0.00 H
| |
| − | HETATM 58 H LIG 1 -3.709 0.878 -0.350 1.00 0.00 H
| |
| − | HETATM 59 H LIG 1 -3.705 0.294 -2.046 1.00 0.00 H
| |
| − | HETATM 60 H LIG 1 -5.261 0.709 -1.249 1.00 0.00 H
| |
| − | HETATM 61 C LIG 1 -4.954 4.653 1.250 1.00 0.00 C
| |
| − | HETATM 62 H LIG 1 -6.034 4.696 0.992 1.00 0.00 H
| |
| − | HETATM 63 H LIG 1 -4.575 5.694 1.319 1.00 0.00 H
| |
| − | HETATM 64 H LIG 1 -4.866 4.160 2.240 1.00 0.00 H
| |
| − | HETATM 65 H LIG 1 3.315 5.492 6.533 1.00 0.00 H
| |
| − | HETATM 66 H LIG 1 4.431 4.561 7.584 1.00 0.00 H
| |
| − | HETATM 67 H LIG 1 2.898 3.803 7.049 1.00 0.00 H
| |
| − | HETATM 68 C LIG 1 -1.636 4.418 4.750 1.00 0.00 C
| |
| − | HETATM 69 C LIG 1 -2.232 5.057 5.855 1.00 0.00 C
| |
| − | HETATM 70 C LIG 1 -1.685 6.253 6.329 1.00 0.00 C
| |
| − | HETATM 71 C LIG 1 -0.594 6.836 5.705 1.00 0.00 C
| |
| − | HETATM 72 C LIG 1 0.008 6.242 4.581 1.00 0.00 C
| |
| − | HETATM 73 H LIG 1 -2.038 3.477 4.395 1.00 0.00 H
| |
| − | HETATM 74 C LIG 1 -3.451 4.513 6.512 1.00 0.00 C
| |
| − | HETATM 75 H LIG 1 -2.125 6.763 7.177 1.00 0.00 H
| |
| − | HETATM 76 H LIG 1 -0.253 7.779 6.099 1.00 0.00 H
| |
| − | HETATM 77 C LIG 1 1.134 6.976 3.886 1.00 0.00 C
| |
| − | HETATM 78 O LIG 1 1.472 6.651 2.760 1.00 0.00 O
| |
| − | HETATM 79 O LIG 1 1.765 8.032 4.452 1.00 0.00 O
| |
| − | HETATM 80 H LIG 1 1.616 8.338 5.353 1.00 0.00 H
| |
| − | HETATM 81 O LIG 1 -3.939 5.126 7.450 1.00 0.00 O
| |
| − | HETATM 82 O LIG 1 -4.098 3.438 5.979 1.00 0.00 O
| |
| − | HETATM 83 N LIG 1 -5.268 2.966 6.406 1.00 0.00 N
| |
| − | HETATM 84 C LIG 1 -6.192 2.486 5.575 1.00 0.00 C
| |
| − | HETATM 85 C LIG 1 -5.710 2.891 7.675 1.00 0.00 C
| |
| − | HETATM 86 O LIG 1 -5.047 3.211 8.649 1.00 0.00 O
| |
| − | HETATM 87 C LIG 1 -7.100 2.339 7.770 1.00 0.00 C
| |
| − | HETATM 88 C LIG 1 -7.441 2.046 6.278 1.00 0.00 C
| |
| − | HETATM 89 H LIG 1 -7.617 0.963 6.107 1.00 0.00 H
| |
| − | HETATM 90 H LIG 1 -8.307 2.651 5.933 1.00 0.00 H
| |
| − | HETATM 91 H LIG 1 -7.781 3.103 8.204 1.00 0.00 H
| |
| − | HETATM 92 H LIG 1 -7.110 1.409 8.375 1.00 0.00 H
| |
| − | HETATM 93 O LIG 1 -6.033 2.434 4.365 1.00 0.00 O
| |
| − | END
| |
| − | </source>
| |
| − | === The testrotkit.pml file ===
| |
| − | <source lang="python">
| |
| − | #-------------------------------------------------------------------------------
| |
| − | # Name: rotkit.py
| |
| − | # Purpose: To rotate molecules easier in pymol
| |
| − | #
| |
| − | # Author: tlinnet
| |
| − | #
| |
| − | # Created: 30/08/2011
| |
| − | # Copyright: (c) tlinnet 2011
| |
| − | # Licence: Free
| |
| − | #-------------------------------------------------------------------------------
| |
| | | | |
| − | cd C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\Distance-Plot
| + | === Functions available in PyMOL === |
| − | import rotkit
| + | * rotateline(Pos1,Pos2,degangle,molecule): |
| | + | *: "Pos1->Pos2" define a line whereabout "molecule" will be rotated "degangle" degrees |
| | + | *: '''rotateline Pos1=P513C_CA, Pos2=P513C_CB, degangle=5, molecule=Atto590''' |
| | + | *: '''rotateline Pos1=dyeatom87, Pos2=dyeatom85, degangle=10, molecule=Atto590''' |
| | + | * mutate(molecule,chain,resi,target="CYS",mutframe="1"): |
| | + | *: Mutate a /molecule//chain/resi into a target, and selecting most probable frame 1 |
| | + | *: '''mutate 1HP1, chain=A, resi=515, target=CYS, mutframe=1''' |
| | + | * toline(Pos1,Pos2,atom,molecule,dist=1): |
| | + | *: Translate molecule atom, 1 angstrom away in the same direction Pos1->Pos2 specify |
| | + | *: '''toline Pos1=P513C_CA, Pos2=P513C_CB, atom=dyeatom87, molecule=Atto590, dist=3''' |
| | | | |
| − | #fetch 1HP1, async=0
| + | '''Available through rotkit.functionname''' |
| − | load 1HP1.pdb
| + | * printMat(matrix): |
| − | load Atto590.pdb
| + | *: prints the TTT matrix in a readable format. (4X4) |
| | + | * getxyz(Sel): |
| | + | *: output is a list [x,y,z] in float. The input can be a list, a string(list) or a selection. |
| | + | * vector(Sel1,Sel2): |
| | + | *: Finds the vector between points. Gets the xyz list from getxyz, so input can be anything. |
| | + | * vectorstr(vector): |
| | + | *: turn a vector in list format into string. No real function actually. |
| | + | * transmat(vector,dist=1): |
| | + | *: Makes a TTT translation matrix for according to the input vector. The vector is multiplied with dist. |
| | + | * unitvector(vector): |
| | + | *: Make a vector a unitvector. |
| | + | * radangle(angle): |
| | + | *: Convert degree to radians. Not that all input are assumed to be in degrees, and are converted automatically. |
| | + | * rotmat(angle,vectornorm,pointcoord): |
| | + | *: This function is the most important. That makes the TTT matrix that rotates a molecule around a normalized vector, which goes through a coordinate point. |
| | + | * crossprod(Vector1, Vector2): |
| | + | *: Makes a crossproduct between two vectors |
| | + | * crosspoint(Pos1, crossprod): |
| | + | *: Returns the endpoint for the Position plus the crossproduct vector. Suitable if one would like to rotate around a crossvector. |
| | | | |
| − | ### Get the names of the loaded objects
| + | <gallery heights="240px" widths="340px"> |
| − | protname = cmd.get_names()[0]
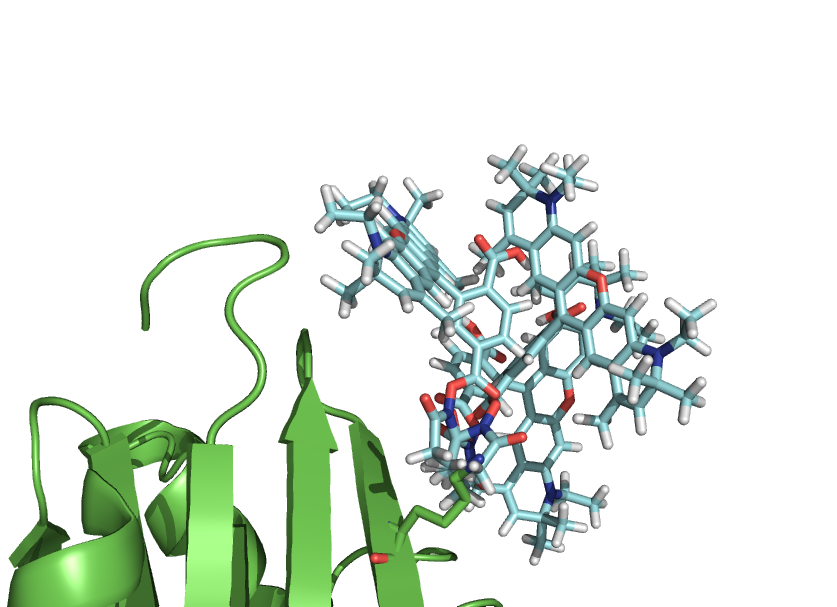
| + | Image:Rotkitex1.png|Place your dye how you want. You always get same ending position. |
| − | molname = cmd.get_names()[1]
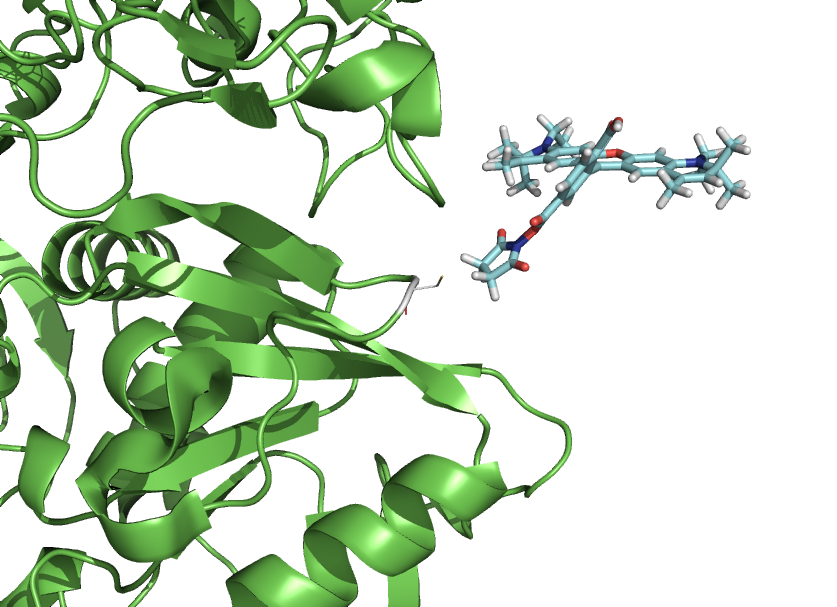
| + | Image:Rotkitex2.png|Mutate a residue through command line, and quickly put your dye there. |
| | + | </gallery> |
| | | | |
| − | ### Make the names we are going to use
| + | == Example of use == |
| − | protselectCB="%s and resi 308 and name CB"%protname
| + | === Example 1 - Make a rotation of domain === |
| − | protnameselectCB="K308CB"
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_1.pml}} |
| − | protselectCA="%s and resi 308 and name CA"%protname
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_1.pml" highlight="python" /> |
| − | protnameselectCA="K308CA"
| |
| − | molselect87="%s and id 87"%molname
| |
| − | molnameselect87="dyeatom87"
| |
| − | molselect85="%s and id 85"%molname
| |
| − | molnameselect85="dyeatom85"
| |
| − | | |
| − | ### Make some selections
| |
| − | cmd.select("%s"%protnameselectCB,"%s"%protselectCB)
| |
| − | cmd.select("%s"%protnameselectCA,"%s"%protselectCA)
| |
| − | cmd.select("%s"%molnameselect85,"%s"%molselect85)
| |
| − | cmd.select("%s"%molnameselect87,"%s"%molselect87)
| |
| − | | |
| − | ### Make nice representations
| |
| − | cmd.show_as("cartoon","%s"%protname)
| |
| − | cmd.show("sticks","byres %s"%protnameselectCB)
| |
| − | | |
| − | ##### PART I: Use of functions #####
| |
| − | ### This view will take you to the first part
| |
| − | set_view (\
| |
| − | 0.377224118, 0.880101919, -0.288305759,\
| |
| − | 0.661396861, -0.473919988, -0.581338286,\
| |
| − | -0.648268998, 0.028612033, -0.760871351,\
| |
| − | 0.000000000, 0.000000000, -56.408561707,\
| |
| − | 19.480533600, 34.572898865, 6.978204727,\
| |
| − | 46.615653992, 66.201446533, -20.000001907 )
| |
| − | | |
| − | #### Just unhash each part for itself, as you continue through
| |
| − | #### The python mini shell is important, to get the return values of the functions
| |
| − | | |
| − | ### To print a objects TTT matrix in a readable format
| |
| − | python
| |
| − | rotkit.printMat(cmd.get_object_matrix(molname))
| |
| − | python end
| |
| − | | |
| − | ##### We want to move the dye to a desired location, and rotate it to a view we desire
| |
| − | ##### First get the vector bewteen the dyeatom and the protein atom
| |
| − | ##python
| |
| − | ##diffvector = rotkit.vector("%s"%molselect87,"%s"%protnameselectCB)
| |
| − | ##python end
| |
| − | ##
| |
| − | ##### Then move the dye
| |
| − | ##python
| |
| − | ##move = rotkit.transmat(diffvector)
| |
| − | ##### print the matrix for fun
| |
| − | ##rotkit.printMat(move)
| |
| − | ##### Move the dye
| |
| − | ##cmd.transform_selection("%s"%molname,move)
| |
| − | ##python end
| |
| − | ##
| |
| − | ##### Now we want to displace the dye in the CA-CB bond direction
| |
| − | ##python
| |
| − | ##### First find the vector/direction to displace it in. From A -> B
| |
| − | ##diffvector = rotkit.vector("%s"%protnameselectCA,"%s"%protnameselectCB)
| |
| − | ##### Make the vector so its lenth is equal 1
| |
| − | ##uvector = rotkit.unitvector(diffvector)[0]
| |
| − | ##### Make the move translation matrix, and we multiply the matrix with 3, so it moves 3 Angstrom
| |
| − | ##move = rotkit.transmat(uvector,3)
| |
| − | ##### Print the matrix
| |
| − | ##rotkit.printMat(move)
| |
| − | ##### Displace it in the CA-CB direction
| |
| − | ##cmd.transform_selection("%s"%molname,move)
| |
| − | ##python end
| |
| − | ##
| |
| − | ##### Now we want to rotate it a single time. We convert 40 degress to radians
| |
| − | ##### The input is the angle, the line to rotate around, and a point where the line goes through
| |
| − | ##python
| |
| − | ##CBxyz = rotkit.getxyz("%s"%protnameselectCB)[0]
| |
| − | ##rmat = rotkit.rotmat(rotkit.radangle(40),uvector,CBxyz)
| |
| − | ##rotkit.printMat(rmat)
| |
| − | ##### Copy paste this line into pymol to see it manually
| |
| − | ##cmd.transform_selection("%s"%molname,rmat)
| |
| − | ##python end
| |
| − | ##
| |
| − | ##### We are not quite satisfied, we want to rotate it around its own bond
| |
| − | ##### So we rotate in around its own 87-85 bonds
| |
| − | ##python
| |
| − | ##diffvector = rotkit.vector("%s"%molnameselect87,"%s"%molnameselect85)
| |
| − | ##uvector = rotkit.unitvector(diffvector)[0]
| |
| − | ##xyz85 = rotkit.getxyz("%s"%molnameselect85)[0]
| |
| − | ##rmat = rotkit.rotmat(rotkit.radangle(10),uvector,xyz85)
| |
| − | ##### Copy paste this line into pymol to see it manually
| |
| − | ##cmd.transform_selection("%s"%molname,rmat)
| |
| − | ##python end
| |
| − | ##
| |
| − | ##### Now, lets make a function that collects all these call in one function
| |
| − | ##### We only want to define two positions that defines the line, the angle and the object to rotate
| |
| − | ##python
| |
| − | ##rotkit.rotateline("%s"%molnameselect87,"%s"%molnameselect85,180,"%s"%molname)
| |
| − | ##python end
| |
| − | ##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
| |
| − | ##print("rotateline Pos1=%s, Pos2=%s, degangle=15, molecule=%s"%(molnameselect87, molnameselect85, molname))
| |
| − | ##
| |
| − | ##### To illustate best, we create som copies of the dye
| |
| − | ##python
| |
| − | ##anglerange = range(90,360,90)
| |
| − | ##for angle in anglerange:
| |
| − | ## ### Make a suitable name for the new molecule
| |
| − | ## molanglename="%s%s"%(molname,angle)
| |
| − | ## ### Now make a copy
| |
| − | ## cmd.create(molanglename,molname)
| |
| − | ## ### Rotate the copy
| |
| − | ## rotkit.rotateline("%s"%molnameselect87,"%s"%molnameselect85,angle,"%s"%molanglename)
| |
| − | ##python end
| |
| − | ##
| |
| − | ####### End of PART I ####
| |
| − | ####### PART II: More advanced functions #####
| |
| − | ##### This view will take you to the second part
| |
| − | ##set_view (\
| |
| − | ## 0.723298192, 0.467510879, 0.508201897,\
| |
| − | ## 0.371686131, -0.883831143, 0.284063697,\
| |
| − | ## 0.581970334, -0.016570913, -0.813038886,\
| |
| − | ## 0.000000000, 0.000000000, -76.609786987,\
| |
| − | ## 11.790571213, 64.992294312, 20.803859711,\
| |
| − | ## -31.181428909, 184.401092529, -20.000001907 )
| |
| − | ##
| |
| − | ##### We can fast mutate a protein. frame 1 is the most probable mutation
| |
| − | ##python
| |
| − | ##rotkit.mutate(protname, chain="A", resi=513, target="CYS", mutframe=1)
| |
| − | ##python end
| |
| − | ##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
| |
| − | ##print("mutate %s, chain=%s, resi=%s, target=CYS, mutframe=1"%(protname, "A", 515))
| |
| − | ##
| |
| − | ##### We now make some selections for this mutation
| |
| − | ##protselectCBcys="%s and resi 513 and name CB"%protname
| |
| − | ##protnameselectCBcys="P513C_CB"
| |
| − | ##protselectCAcys="%s and resi 513 and name CA"%protname
| |
| − | ##protnameselectCAcys="P513C_CA"
| |
| − | ##cmd.select("%s"%protnameselectCBcys,"%s"%protselectCBcys)
| |
| − | ##cmd.select("%s"%protnameselectCAcys,"%s"%protselectCAcys)
| |
| − | ##
| |
| − | ##### Now, lets make a function that collects all the commands to put on an atom on the same line defined by two points
| |
| − | ##### The input is the two points that define the line, the atom of a molecule to be put on the line, and the distance to move
| |
| − | ##python
| |
| − | ##rotkit.toline(protnameselectCAcys,protnameselectCBcys,molnameselect87,molname,3)
| |
| − | ##rotkit.rotateline(protnameselectCAcys,protnameselectCBcys,5,molname)
| |
| − | ##rotkit.rotateline(molnameselect87,molnameselect85,10,molname)
| |
| − | ##python end
| |
| − | ##print("toline Pos1=%s, Pos2=%s, atom=%s, molecule=%s, dist=%s"%(protnameselectCAcys,protnameselectCBcys,molnameselect87,molname,3))
| |
| − | ##print("rotateline Pos1=%s, Pos2=%s, degangle=5, molecule=%s"%(protnameselectCAcys, protnameselectCBcys, molname))
| |
| − | ##print("rotateline Pos1=%s, Pos2=%s, degangle=10, molecule=%s"%(molnameselect87, molnameselect85, molname))
| |
| − | ##
| |
| − | ####### End of PART II ####
| |
| − | </source>
| |
| − | | |
| − | == rotkit.py ==
| |
| − | The code can be downloaded fast from here http://tinyurl.com/pymolrotkit <br />
| |
| − | # wget http://tinyurl.com/pymolrotkit
| |
| − | # mv pymolrotkit rotkit.py
| |
| − | | |
| − | <source lang="python">
| |
| − | #-------------------------------------------------------------------------------
| |
| − | # Name: rotkit.py examples
| |
| − | # Purpose: To rotate molecules easier
| |
| − | #
| |
| − | # Author: tlinnet
| |
| − | #
| |
| − | # Created: 30/08/2011
| |
| − | # Copyright: (c) tlinnet 2011
| |
| − | # Licence: Free
| |
| − | #-------------------------------------------------------------------------------
| |
| − | | |
| − | | |
| − | from pymol import cmd
| |
| − | import math
| |
| − | | |
| − | def printMat(matrix):
| |
| − | print("%s %s %s %s \n%s %s %s %s \n%s %s %s %s \n%s %s %s %s"%(matrix[0],matrix[1],matrix[2],matrix[3],matrix[4],matrix[5],matrix[6],matrix[7],matrix[8],matrix[9],matrix[10],matrix[11],matrix[12],matrix[13],matrix[14],matrix[15]))
| |
| − | return None
| |
| − | | |
| − | def getxyz(Sel):
| |
| − | if type(Sel)==list and len(Sel)==3:
| |
| − | return Sel, "listXYZ"
| |
| − | if type(Sel)==str and Sel[0]=="[" and Sel[-1]=="]":
| |
| − | Selsplit = list(Sel[1:-1].split(","))
| |
| − | Selsplit = [float(x) for x in Selsplit]
| |
| − | return Selsplit, "strXYZ"
| |
| − | if type(Sel)==str:
| |
| − | pos = cmd.get_atom_coords(Sel)
| |
| − | return pos, "selXYZ"
| |
| − | | |
| − | def vector(Sel1,Sel2):
| |
| − | PosSel1 = getxyz(Sel1)[0]
| |
| − | PosSel2 = getxyz(Sel2)[0]
| |
| − | vectorcalc = [PosSel2[0]-PosSel1[0],PosSel2[1]-PosSel1[1],PosSel2[2]-PosSel1[2]]
| |
| − | return(vectorcalc)
| |
| − | | |
| − | def vectorstr(vector):
| |
| − | return("[%s,%s,%s]"%(vector[0],vector[1],vector[2]))
| |
| − | | |
| − | def transmat(vector,dist=1):
| |
| − | mat = [1,0,0,0,0,1,0,0,0,0,1,0,dist*vector[0],dist*vector[1],dist*vector[2],1]
| |
| − | return(mat)
| |
| − | | |
| − | def unitvector(vector):
| |
| − | vectorlen = math.sqrt(math.pow(vector[0],2)+math.pow(vector[1],2)+math.pow(vector[2],2))
| |
| − | vectordiv = [vector[0]/vectorlen, vector[1]/vectorlen, vector[2]/vectorlen]
| |
| − | return(vectordiv,vectorlen)
| |
| − | | |
| − | def radangle(angle):
| |
| − | return(math.radians(angle))
| |
| − | | |
| − | def rotmat(angle,vectornorm,pointcoord):
| |
| − | ### From: http://inside.mines.edu/~gmurray/ArbitraryAxisRotation/ Section 6.2
| |
| − | u,v,w = vectornorm
| |
| − | a,b,c = pointcoord
| |
| − | makerotmat = [(math.pow(u,2)+(math.pow(v,2)+math.pow(w,2))*math.cos(angle)),
| |
| − | (u*v*(1-math.cos(angle))-w*math.sin(angle)),
| |
| − | (u*w*(1-math.cos(angle))+v*math.sin(angle)),
| |
| − | ((a*(math.pow(v,2)+math.pow(w,2))-u*(b*v+c*w))*(1-math.cos(angle))+(b*w-c*v)*math.sin(angle)),
| |
| − | (u*v*(1-math.cos(angle))+w*math.sin(angle)),
| |
| − | (math.pow(v,2)+(math.pow(u,2)+math.pow(w,2))*math.cos(angle)),
| |
| − | (v*w*(1-math.cos(angle))-u*math.sin(angle)),
| |
| − | ((b*(math.pow(u,2)+math.pow(w,2))-v*(a*u+c*w))*(1-math.cos(angle))+(c*u-a*w)*math.sin(angle)),
| |
| − | (u*w*(1-math.cos(angle))-v*math.sin(angle)),
| |
| − | (v*w*(1-math.cos(angle))+u*math.sin(angle)),
| |
| − | (math.pow(w,2)+(math.pow(u,2)+math.pow(v,2))*math.cos(angle)),
| |
| − | ((c*(math.pow(u,2)+math.pow(v,2))-w*(a*u+b*v))*(1-math.cos(angle))+(a*v-b*u)*math.sin(angle)),
| |
| − | (0),(0),(0),(1),]
| |
| − | return(makerotmat)
| |
| | | | |
| − | def rotateline(Pos1,Pos2,degangle,molecule):
| + | === Example 2 - Simulate dye freedom === |
| − | diffvector = vector(Pos1,Pos2)
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_2.pml}} |
| − | uvector = unitvector(diffvector)[0]
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_2.pml" highlight="python" /> |
| − | xyz = getxyz(Pos2)[0]
| |
| − | rmat = rotmat(radangle(float(degangle)),uvector,xyz)
| |
| − | cmd.transform_selection(molecule,rmat)
| |
| − | return(None)
| |
| − | cmd.extend("rotateline",rotateline)
| |
| | | | |
| − | def mutate(molecule,chain,resi,target="CYS",mutframe="1"):
| + | === Example 3 - Create distance distribution histogram === |
| − | target = target.upper()
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_3.pml}} |
| − | cmd.wizard("mutagenesis")
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_3.pml" highlight="python" /> |
| − | cmd.do("refresh_wizard")
| |
| − | cmd.get_wizard().set_mode("%s"%target)
| |
| − | selection="/%s//%s/%s"%(molecule,chain,resi)
| |
| − | cmd.get_wizard().do_select(selection)
| |
| − | cmd.frame(str(mutframe))
| |
| − | cmd.get_wizard().apply()
| |
| − | cmd.set_wizard("done")
| |
| − | cmd.select("%s%s%s_%s"%(molecule,chain,resi,target),"byres %s"%(selection))
| |
| − | cmd.extend("mutate",mutate)
| |
| | | | |
| − | def toline(Pos1,Pos2,atom,molecule,dist=1):
| + | === Example 4 - A tutorial file === |
| − | dist = float(dist)
| + | To understand how the functions works, read through the tutorial. Hash/Unhash "##" each step at the time to see the effect. |
| − | diffvector = vector(atom,Pos2)
| + | To be able to follow the tutorial, you need the dye molecule, which is loaded from the Pymol-script-repository. |
| − | move = transmat(diffvector)
| |
| − | cmd.transform_selection("%s"%molecule,move)
| |
| − | diffvector = vector(Pos1,Pos2)
| |
| − | uvector = unitvector(diffvector)[0]
| |
| − | move = transmat(uvector,dist)
| |
| − | cmd.transform_selection("%s"%molecule,move)
| |
| − | return(None)
| |
| − | cmd.extend("toline",toline)
| |
| − | </source>
| |
| | | | |
| | + | {{Template:PymolScriptRepoDownload|examples/rotkit_4.pml}} |
| | + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_4.pml" highlight="python" /> |
| | | | |
| | [[Category:Script_Library]] | | [[Category:Script_Library]] |
| | [[Category:ObjSel_Scripts]] | | [[Category:ObjSel_Scripts]] |
| | + | [[Category:Pymol-script-repo]] |