This is a read-only mirror of pymolwiki.org
Difference between revisions of "Rotkit"
| Line 218: | Line 218: | ||
cmd.refresh() | cmd.refresh() | ||
| + | # Make a distribution for the Open case | ||
Don_Acc_distribution = [] | Don_Acc_distribution = [] | ||
python | python | ||
| Line 227: | Line 228: | ||
cmd.delete(distname) | cmd.delete(distname) | ||
python end | python end | ||
| − | Newdir=rotkit.createdirs() | + | Newdir=rotkit.createdirs("results_rotkit") |
os.chdir(Newdir) | os.chdir(Newdir) | ||
| − | rotkit.makehistogram(Don_Acc_distribution,dataname=" | + | rotkit.makehistogram(Don_Acc_distribution,dataname="Don_Acc_Open",datalistindex=2,nrbins=100,binrange=[0,0]) |
| + | |||
| + | # Make a distribution for angle range | ||
| + | cmd.create("Acceptor_rot","All_Acceptors") | ||
| + | python | ||
| + | ang_incr = 1 | ||
| + | anglerange = range(2,98,ang_incr) | ||
| + | nrstates = len(anglerange)+1 | ||
| + | states = 1 | ||
| + | for angle in anglerange: | ||
| + | states += 1 | ||
| + | rot_Acceptor = "Acceptor_rot_%s"%angle | ||
| + | cmd.create(rot_Acceptor,"Acceptor_rot") | ||
| + | rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",-(angle-1),rot_Acceptor) | ||
| + | cmd.create("Acceptor_rot",rot_Acceptor,1,states) | ||
| + | cmd.create("Acceptor_rot",rot_Acceptor,1,2*nrstates-states) | ||
| + | cmd.delete(rot_Acceptor) | ||
| + | for Acc in Acceptor_names: | ||
| + | rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",(angle-1),Acc[1]) | ||
| + | python end | ||
</syntaxhighlight> | </syntaxhighlight> | ||
Revision as of 20:39, 2 December 2011
Author
This pymol script is made by Troels Emtekær Linnet
Introduction
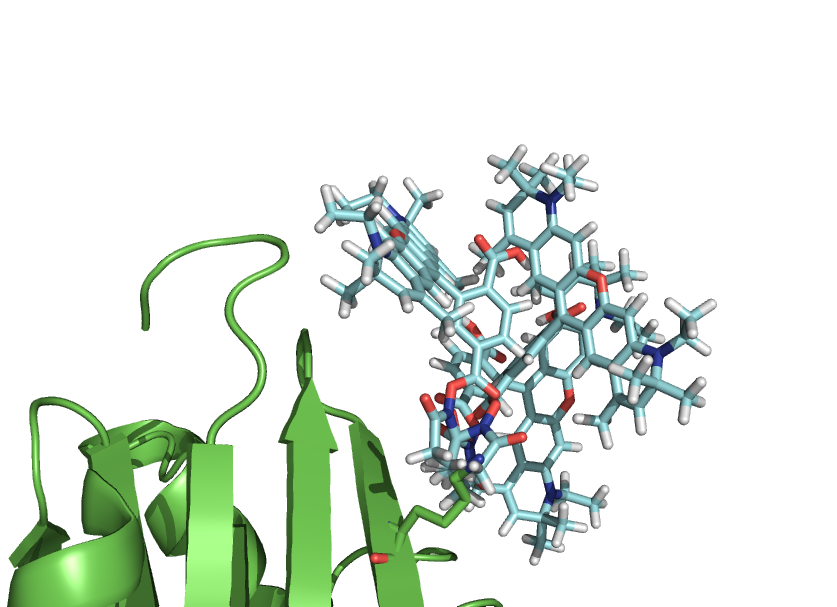
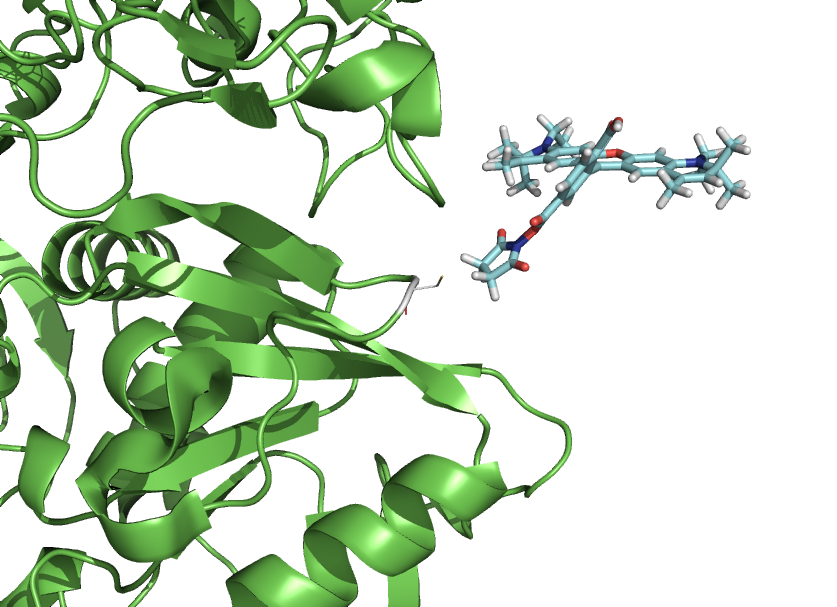
This script-kit is a collection of small script to be able to precisely to put a molecule (like a dye) where you want in relation to a protein.
You can also create rotational states of a domain or simulate a dye freedom.
It simply makes the PyMOL TTT matrixes, in a easy and user friendly way. The calls to the functions available in PyMOL, takes care of all the conversion of input and such.
If you are interested in this, you might also want to check out the PyMOL Chempy module that is included in PyMOL. It provides handy vector and matrix functions.
Functions available in PyMOL
- rotateline(Pos1,Pos2,degangle,molecule):
- "Pos1->Pos2" define a line whereabout "molecule" will be rotated "degangle" degrees
- rotateline Pos1=P513C_CA, Pos2=P513C_CB, degangle=5, molecule=Atto590
- rotateline Pos1=dyeatom87, Pos2=dyeatom85, degangle=10, molecule=Atto590
- mutate(molecule,chain,resi,target="CYS",mutframe="1"):
- Mutate a /molecule//chain/resi into a target, and selecting most probable frame 1
- mutate 1HP1, chain=A, resi=515, target=CYS, mutframe=1
- toline(Pos1,Pos2,atom,molecule,dist=1):
- Translate molecule atom, 1 angstrom away in the same direction Pos1->Pos2 specify
- toline Pos1=P513C_CA, Pos2=P513C_CB, atom=dyeatom87, molecule=Atto590, dist=3
Available through rotkit.functionname
- printMat(matrix):
- prints the TTT matrix in a readable format. (4X4)
- getxyz(Sel):
- output is a list [x,y,z] in float. The input can be a list, a string(list) or a selection.
- vector(Sel1,Sel2):
- Finds the vector between points. Gets the xyz list from getxyz, so input can be anything.
- vectorstr(vector):
- turn a vector in list format into string. No real function actually.
- transmat(vector,dist=1):
- Makes a TTT translation matrix for according to the input vector. The vector is multiplied with dist.
- unitvector(vector):
- Make a vector a unitvector.
- radangle(angle):
- Convert degree to radians. Not that all input are assumed to be in degrees, and are converted automatically.
- rotmat(angle,vectornorm,pointcoord):
- This function is the most important. That makes the TTT matrix that rotates a molecule around a normalized vector, which goes through a coordinate point.
- crossprod(Vector1, Vector2):
- Makes a crossproduct between two vectors
- crosspoint(Pos1, crossprod):
- Returns the endpoint for the Position plus the crossproduct vector. Suitable if one would like to rotate around a crossvector.
Example of use
Make a rotation of domain
reinitialize
import rotkit
fetch 1HP1, async=0
show_as cartoon, 1HP1
show_as sticks, 1HP1 and resn ATP
###################### Make rotation axis #################
pseudoatom axisA, vdw=1.0
pseudoatom axisB, vdw=1.0
rotkit.toline("/1HP1//A/477/C","/1HP1//A/423/CG1","axisA","axisA",20)
rotkit.toline("/1HP1//A/423/CG1","/1HP1//A/477/C","axisB","axisB",5)
show spheres, axisA or axisB
label axisA, "axisA"
label axisB, "axisB"
dist rotaxis, axisA, axisB
color green, rotaxis
set dash_width, 5
set dash_gap, 0
hide label, rotaxis
## Create rotate states of 1HP1
create 1HP1_rot, 1HP1, 1, 1
python
ang_incr = 1
anglerange = range(2,98,ang_incr)
nrstates = len(anglerange)+1
states = 1
for angle in anglerange:
states += 1
rot_1HP1 = "1HP1_rot_%s"%angle
cmd.create(rot_1HP1,"(1HP1 and resi 363-550) or (1HP1 and resn ATP)")
rotkit.rotateline("axisA","axisB",-(angle-1),rot_1HP1)
cmd.create("1HP1_rot",rot_1HP1,1,states)
cmd.create("1HP1_rot",rot_1HP1,1,2*nrstates-states)
cmd.delete(rot_1HP1)
python end
hide cartoon, (1HP1 and resi 363-550)
hide sticks, (1HP1 and resn ATP)
mplay
Simulate dye freedom
reinitialize
import rotkit
fetch 1HP1, async=0
show_as cartoon, 1HP1
show_as sticks, 1HP1 and resn ATP
set auto_zoom, off
###################### Make rotation axis #################
pseudoatom axisA, vdw=1.0
pseudoatom axisB, vdw=1.0
rotkit.toline("/1HP1//A/477/C","/1HP1//A/423/CG1","axisA","axisA",20)
rotkit.toline("/1HP1//A/423/CG1","/1HP1//A/477/C","axisB","axisB",5)
show spheres, axisA or axisB
label axisA, "axisA"
label axisB, "axisB"
dist rotaxis, axisA, axisB
color green, rotaxis
set dash_width, 5
set dash_gap, 0
hide label, rotaxis
####################### Create rotate states of dye atoms ###################
##### First mutate, the mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
rotkit.mutate("1HP1", chain="A", resi=308, target="CYS", mutframe=1)
cmd.refresh()
rotkit.mutate("1HP1", chain="A", resi=513, target="CYS", mutframe=1)
cmd.refresh()
##### Create simulated dye movement atoms
pseudoatom Donor, vdw=0.5
pseudoatom Acceptor, vdw=0.5
show spheres, Donor or Acceptor
rotkit.toline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name SG","Donor","Donor",15.0)
rotkit.toline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name SG","Acceptor","Acceptor",15.0)
python
Dye_ang_incr = 6
Donor_angle_range = range(0,359,Dye_ang_incr)
Acceptor_angle_range = range(0,359,Dye_ang_incr)
nrstates = len(Donor_angle_range)+1
Donor_states = 1
Acceptor_states = 1
for Donor_angle in Donor_angle_range:
Donor_states += 1
Donor_angle_name="Donor_%s"%(Donor_angle)
cmd.create(Donor_angle_name,"Donor")
rotkit.rotateline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name CB",Donor_angle,Donor_angle_name)
# Save it as states in Donor
cmd.create("Donor",Donor_angle_name,1,Donor_states)
cmd.create("Donor",Donor_angle_name,1,2*nrstates-Donor_states)
cmd.group("All_Donors",Donor_angle_name)
for Acceptor_angle in Acceptor_angle_range:
Acceptor_states += 1
Acceptor_angle_name="Acceptor_%s"%(Acceptor_angle)
cmd.create(Acceptor_angle_name,"Acceptor")
rotkit.rotateline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name CB",Acceptor_angle,Acceptor_angle_name)
# Save it as states in Acceptor
cmd.create("Acceptor",Acceptor_angle_name,1,Acceptor_states)
cmd.create("Acceptor",Acceptor_angle_name,1,2*nrstates-Acceptor_states)
cmd.group("All_Acceptors",Acceptor_angle_name)
python end
disable All_Donors
disable All_Acceptors
cmd.create("Donor","All_Donors",1,1)
cmd.create("Acceptor","All_Acceptors",1,1)
mplay
Create distance distribution histogram
reinitialize
#You need to make sure you are in the right dir, since we are going to make some datafiles
cd /home/tlinnet/test
import rotkit
fetch 1HP1, async=0
show_as cartoon, 1HP1
show_as sticks, 1HP1 and resn ATP
set auto_zoom, off
####################### Create rotate states of dye atoms ###################
##### First mutate, the mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
rotkit.mutate("1HP1", chain="A", resi=308, target="CYS", mutframe=1)
cmd.refresh()
rotkit.mutate("1HP1", chain="A", resi=513, target="CYS", mutframe=1)
cmd.refresh()
##### Create simulated dye movement atoms
pseudoatom Donor, vdw=0.5
pseudoatom Acceptor, vdw=0.5
show spheres, Donor or Acceptor
rotkit.toline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name SG","Donor","Donor",15.0)
rotkit.toline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name SG","Acceptor","Acceptor",15.0)
python
Dye_ang_incr = 6
Donor_angle_range = range(0,359,Dye_ang_incr)
Acceptor_angle_range = range(0,359,Dye_ang_incr)
Donor_names = []
Acceptor_names = []
for Donor_angle in Donor_angle_range:
Donor_angle_name="Donor_%s"%(Donor_angle)
Donor_names.append([Donor_angle,Donor_angle_name])
cmd.create(Donor_angle_name,"Donor")
rotkit.rotateline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name CB",Donor_angle,Donor_angle_name)
cmd.group("All_Donors",Donor_angle_name)
for Acceptor_angle in Acceptor_angle_range:
Acceptor_angle_name="Acceptor_%s"%(Acceptor_angle)
Acceptor_names.append([Acceptor_angle,Acceptor_angle_name])
cmd.create(Acceptor_angle_name,"Acceptor")
rotkit.rotateline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name CB",Acceptor_angle,Acceptor_angle_name)
cmd.group("All_Acceptors",Acceptor_angle_name)
python end
disable All_Donors
disable All_Acceptors
cmd.create("Donor","All_Donors")
cmd.create("Acceptor","All_Acceptors")
cmd.refresh()
# Make a distribution for the Open case
Don_Acc_distribution = []
python
for Don in Donor_names:
for Acc in Acceptor_names:
distname = "%s_%s"%(Don[1],Acc[1])
distance = cmd.dist(distname,Don[1],Acc[1])
Don_Acc_distribution.append([Don[0], Acc[1], distance])
cmd.delete(distname)
python end
Newdir=rotkit.createdirs("results_rotkit")
os.chdir(Newdir)
rotkit.makehistogram(Don_Acc_distribution,dataname="Don_Acc_Open",datalistindex=2,nrbins=100,binrange=[0,0])
# Make a distribution for angle range
cmd.create("Acceptor_rot","All_Acceptors")
python
ang_incr = 1
anglerange = range(2,98,ang_incr)
nrstates = len(anglerange)+1
states = 1
for angle in anglerange:
states += 1
rot_Acceptor = "Acceptor_rot_%s"%angle
cmd.create(rot_Acceptor,"Acceptor_rot")
rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",-(angle-1),rot_Acceptor)
cmd.create("Acceptor_rot",rot_Acceptor,1,states)
cmd.create("Acceptor_rot",rot_Acceptor,1,2*nrstates-states)
cmd.delete(rot_Acceptor)
for Acc in Acceptor_names:
rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",(angle-1),Acc[1])
python end
A tutorial.pml file
To understand how the functions works, read through the tutorial. Hash/Unhash "##" each step at the time to see the effect.
Make a test dir: /home/tlinnet/test and make a tutorial.pml with the commands.
To be able to follow the tutorial, you need the dye molecule. Right clicking the following link here -> Save as: Atto590.pdb in the directory with the tutorial.pml file
https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/files_for_examples/Atto590.pdb
reinitialize
cd /home/tlinnet/test
import rotkit
fetch 1HP1, async=0
load Atto590.pdb
# Make sure everything is loaded before we continue
cmd.refresh()
### Get the names of the loaded objects
protname = cmd.get_names()[0]
molname = cmd.get_names()[1]
### Make the names we are going to use
protselectCB="%s and resi 308 and name CB"%protname
protnameselectCB="K308CB"
protselectCA="%s and resi 308 and name CA"%protname
protnameselectCA="K308CA"
molselect13="%s and id 13"%molname
molnameselect13="dyeatom13"
molselect12="%s and id 12"%molname
molnameselect12="dyeatom12"
### Make some selections
cmd.select("%s"%protnameselectCB,"%s"%protselectCB)
cmd.select("%s"%protnameselectCA,"%s"%protselectCA)
cmd.select("%s"%molnameselect13,"%s"%molselect13)
cmd.label("%s"%molnameselect13,"13")
cmd.select("%s"%molnameselect12,"%s"%molselect12)
cmd.label("%s"%molnameselect12,"12")
### Make nice representations
cmd.show_as("cartoon","%s"%protname)
cmd.show("sticks","byres %s"%protnameselectCB)
##### PART I: Use of functions #####
### This view will take you to the first part
set_view (\
0.377224118, 0.880101919, -0.288305759,\
0.661396861, -0.473919988, -0.581338286,\
-0.648268998, 0.028612033, -0.760871351,\
0.000000000, 0.000000000, -56.408561707,\
19.480533600, 34.572898865, 6.978204727,\
46.615653992, 66.201446533, -20.000001907 )
#### Just unhash each part for itself, as you continue through
### To print a objects TTT matrix in a readable format
rotkit.printMat(cmd.get_object_matrix(molname))
##### We want to move the dye to a desired location, and rotate it to a view we desire
##### First get the vector bewteen the dyeatom and the protein atom
diffvector = rotkit.vector("%s"%molselect13,"%s"%protnameselectCB)
##### Then move the dye
move = rotkit.transmat(diffvector)
##### print the matrix for fun
rotkit.printMat(move)
##### Move the dye
cmd.transform_selection("%s"%molname,move)
##### Now we want to displace the dye in the CA-CB bond direction
##### First find the vector/direction to displace it in. From A -> B
diffvector = rotkit.vector("%s"%protnameselectCA,"%s"%protnameselectCB)
##### Make the vector so its lenth is equal 1
uvector = rotkit.unitvector(diffvector)[0]
##### Make the move translation matrix, and we multiply the matrix with 3, so it moves 3 Angstrom
move = rotkit.transmat(uvector,3)
##### Print the matrix
rotkit.printMat(move)
##### Displace it in the CA-CB direction
cmd.transform_selection("%s"%molname,move)
##### Now we want to rotate it a single time. We convert 40 degress to radians
##### The input is the angle, the line to rotate around, and a point where the line goes through
CBxyz = rotkit.getxyz("%s"%protnameselectCB)[0]
rmat = rotkit.rotmat(rotkit.radangle(40),uvector,CBxyz)
rotkit.printMat(rmat)
##### Copy paste this line into pymol to see it manually
cmd.transform_selection("%s"%molname,rmat)
##### We are not quite satisfied, we want to rotate it around its own bond
##### So we rotate in around its own 13 -> 12 bonds
diffvector = rotkit.vector("%s"%molnameselect13,"%s"%molnameselect12)
uvector = rotkit.unitvector(diffvector)[0]
xyz12 = rotkit.getxyz("%s"%molnameselect12)[0]
rmat = rotkit.rotmat(rotkit.radangle(10),uvector,xyz12)
##### Copy paste this line into pymol to see it manually
cmd.transform_selection("%s"%molname,rmat)
##### Now, lets make a function that collects all these call in one function
##### We only want to define two positions that defines the line, the angle and the object to rotate
rotkit.rotateline("%s"%molnameselect13,"%s"%molnameselect12,180,"%s"%molname)
##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
print("rotateline Pos1=%s, Pos2=%s, degangle=15, molecule=%s"%(molnameselect13, molnameselect12, molname))
##### To illustate best, we create som copies of the dye
python
anglerange = range(90,360,90)
for angle in anglerange:
### Make a suitable name for the new molecule
molanglename="%s%s"%(molname,angle)
### Now make a copy
cmd.create(molanglename,molname)
cmd.label("%s and id 12"%molanglename,"12")
cmd.label("%s and id 13"%molanglename,"13")
### Rotate the copy
rotkit.rotateline("%s"%protnameselectCB,"%s"%molnameselect13,angle,"%s"%molanglename)
python end
####### End of PART I ####
####### PART II: More advanced functions #####
##### This view will take you to the second part
set_view (\
0.723298192, 0.467510879, 0.508201897,\
0.371686131, -0.883831143, 0.284063697,\
0.581970334, -0.016570913, -0.813038886,\
0.000000000, 0.000000000, -76.609786987,\
11.790571213, 64.992294312, 20.803859711,\
-31.181428909, 184.401092529, -20.000001907 )
##### We can fast mutate a protein. frame 1 is the most probable mutation
rotkit.mutate(protname, chain="A", resi=513, target="CYS", mutframe=1)
##### The mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
cmd.refresh()
##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
print("mutate %s, chain=%s, resi=%s, target=CYS, mutframe=1"%(protname, "A", 515))
##### We now make some selections for this mutation
protselectCBcys="%s and resi 513 and name CB"%protname
protnameselectCBcys="P513C_CB"
protselectCAcys="%s and resi 513 and name CA"%protname
protnameselectCAcys="P513C_CA"
cmd.select("%s"%protnameselectCBcys,"%s"%protselectCBcys)
cmd.select("%s"%protnameselectCAcys,"%s"%protselectCAcys)
##### Now, lets make a function that collects all the commands to put on an atom on the same line defined by two points
##### The input is the two points that define the line, the atom of a molecule to be put on the line, and the distance to move
rotkit.toline(protnameselectCAcys,protnameselectCBcys,molnameselect13,molname,3)
rotkit.rotateline(protnameselectCAcys,protnameselectCBcys,180,molname)
rotkit.rotateline(molnameselect13,molnameselect12,10,molname)
print("toline Pos1=%s, Pos2=%s, atom=%s, molecule=%s, dist=%s"%(protnameselectCAcys,protnameselectCBcys,molnameselect13,molname,3))
print("rotateline Pos1=%s, Pos2=%s, degangle=180, molecule=%s"%(protnameselectCAcys, protnameselectCBcys, molname))
print("rotateline Pos1=%s, Pos2=%s, degangle=10, molecule=%s"%(molnameselect13, molnameselect12, molname))
cmd.refresh()
####### End of PART II ####
####### Now we make a cross product ####
molselect14="%s and id 14"%molname
molnameselect14="dyeatom14"
cmd.select("%s"%molnameselect14,"%s"%molselect14)
cmd.label("%s"%molnameselect14,"14")
cross = rotkit.crossprod(rotkit.vector(molselect13,molselect12),rotkit.vector(molselect13,molselect14))
unity_cross = rotkit.unitvector(cross)[0]
point_cross = rotkit.crosspoint(molselect13,cross)
rotkit.rotateline(molnameselect13,point_cross,180,molname)
print("rotateline Pos1=%s, Pos2=%s, degangle=10, molecule=%s"%(molnameselect13, pcross, molname))
Python Code
This code has been put under version control. In the project, Pymol-script-repo.
For a color coded view:
https://github.com/Pymol-Scripts/Pymol-script-repo/blob/master/rotkit.py
See the raw code or download manually, by right clicking the following link here -> Save as: rotkit.py
https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/rotkit.py