This is a read-only mirror of pymolwiki.org
Difference between revisions of "PoseView"
Jump to navigation
Jump to search
m (1 revision) |
m (1 revision) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 21: | Line 21: | ||
== Usage == | == Usage == | ||
| − | poseview [ ligand [, protein [, width [, height [, exe [, state ]]]]]] | + | poseview [ ligand [, protein [, width [, height [, filename [, exe [, state ]]]]]]] |
== Example == | == Example == | ||
Latest revision as of 04:20, 4 February 2019
| Type | Python Module |
|---|---|
| Download | poseview.py |
| Author(s) | Thomas Holder |
| License | BSD-2-Clause |
| This code has been put under version control in the project Pymol-script-repo | |
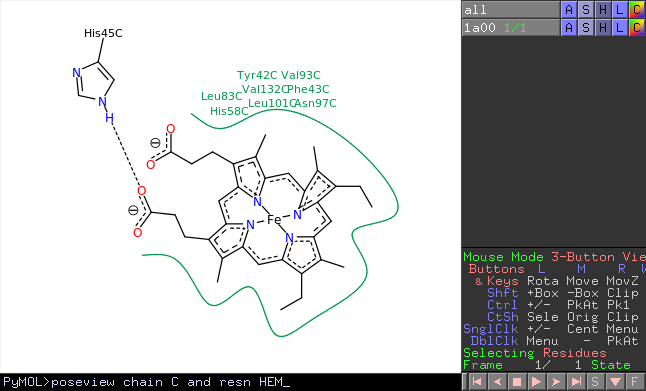
poseview is a PyMOL wrapper for the PoseView program. PoseView generates 2D structure-diagrams of protein-ligand complexes.
Setup
You need the poseview executable and a valid license file. Example for a launcher script which should be saved to /usr/bin/poseview so that PyMOL can find it:
#!/bin/sh
POSEVIEW_PATH="/opt/poseview-1.1.2-Linux-x64"
export BIOSOLVE_LICENSE_FILE="$POSEVIEW_PATH/poseview.lic"
exec "$POSEVIEW_PATH/poseview" "$@"
Usage
poseview [ ligand [, protein [, width [, height [, filename [, exe [, state ]]]]]]]
Example
fetch 1a00, async=0
poseview chain C and resn HEM