This is a read-only mirror of pymolwiki.org
FindSurfaceResidues
Jump to navigation
Jump to search
| Type | Python Module |
|---|---|
| Download | findSurfaceResidues.py |
| Author(s) | Jason Vertrees |
| License | BSD-2-Clause |
| This code has been put under version control in the project Pymol-script-repo | |
Overview
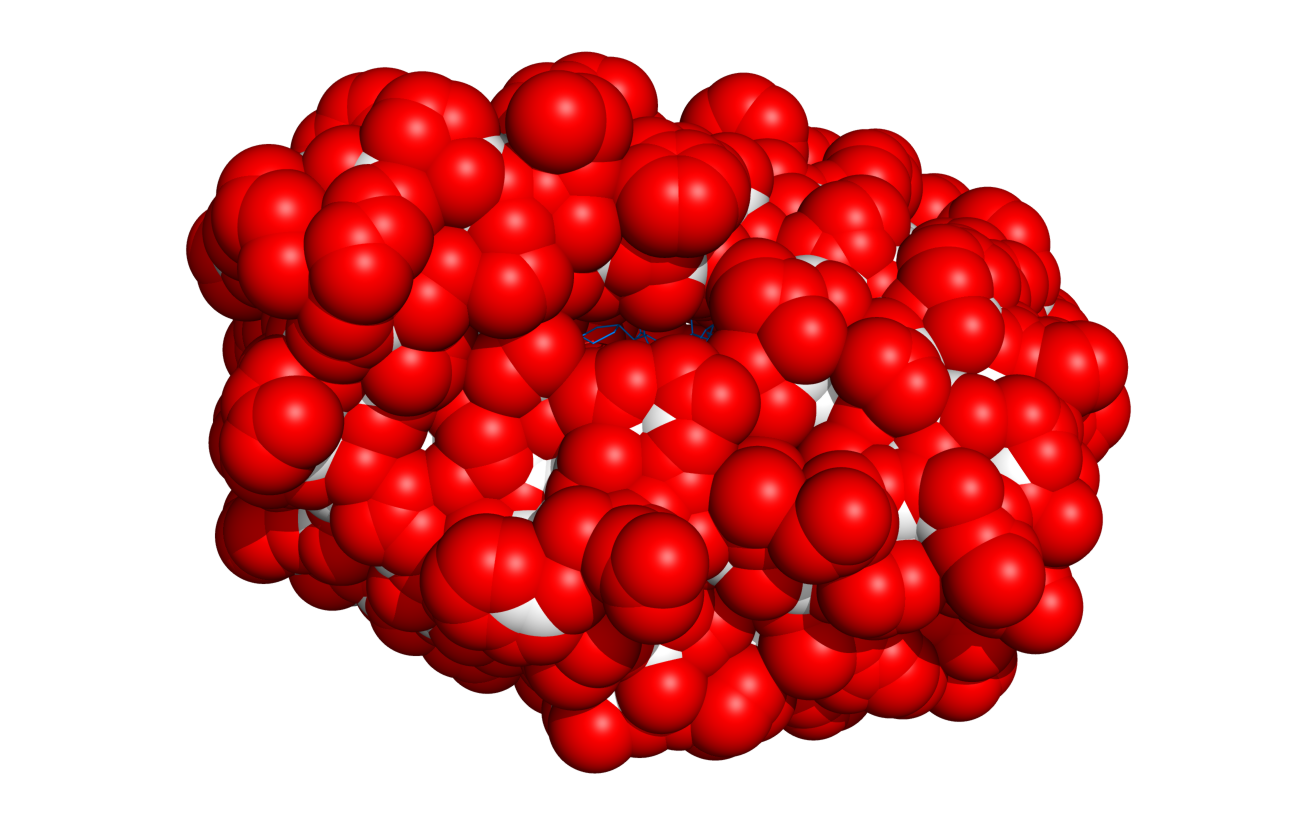
This script will select (and color if requested) surface residues and atoms on an object or selection. See the options below.
Each time, the script will create two new selections called, 'exposed_res_XYZ' and 'exposed_atm_XYZ' where XYZ is some random number. This is done so that no other selections/objects are overwritten.
Usage
findSurfaceResidues [ selection=all [, cutoff=2.5 [, doShow=False ]]]
The parameters are:
selection
- The object or selection for which to find exposed residues;
- DEFAULT = (all)
cutoff
- The cutoff in square Angstroms that defines exposed or not. Those residues with > cutoff Ang^2 exposed will be considered exposed;
- DEFAULT = 2.5 Ang^2
doShow
- Change the visualization to highlight the exposed residues vs interior
- DEFAULT = False/Blank
Examples
# make sure you download and run the code below, before trying these examples.
load $TUT/1hpv.pdb
findSurfaceResidues
# now show the exposed
findSurfaceResidues doShow=True
# watch how the visualization changes:
findSurfaceResidues doShow=1, cutoff=0.5
findSurfaceResidues doShow=1, cutoff=1.0
findSurfaceResidues doShow=1, cutoff=1.5
findSurfaceResidues doShow=1, cutoff=2.0
findSurfaceResidues doShow=1, cutoff=2.5
findSurfaceResidues doShow=1, cutoff=3.0
The Code
# -*- coding: utf-8 -*-
import pymol
from pymol import cmd
import random
def findSurfaceResidues(objSel="(all)", cutoff=2.5, doShow=False, verbose=False):
"""
findSurfaceResidues
finds those residues on the surface of a protein
that have at least 'cutoff' exposed A**2 surface area.
PARAMS
objSel (string)
the object or selection in which to find
exposed residues
DEFAULT: (all)
cutoff (float)
your cutoff of what is exposed or not.
DEFAULT: 2.5 Ang**2
asSel (boolean)
make a selection out of the residues found
RETURNS
(list: (chain, resv ) )
A Python list of residue numbers corresponding
to those residues w/more exposure than the cutoff.
"""
tmpObj="__tmp"
cmd.create( tmpObj, objSel + " and polymer");
if verbose!=False:
print "WARNING: I'm setting dot_solvent. You may not care for this."
cmd.set("dot_solvent");
cmd.get_area(selection=tmpObj, load_b=1)
# threshold on what one considers an "exposed" atom (in A**2):
cmd.remove( tmpObj + " and b < " + str(cutoff) )
stored.tmp_dict = {}
cmd.iterate(tmpObj, "stored.tmp_dict[(chain,resv)]=1")
exposed = stored.tmp_dict.keys()
exposed.sort()
randstr = str(random.randint(0,10000))
selName = "exposed_atm_" + randstr
if verbose!=False:
print "Exposed residues are selected in: " + selName
cmd.select(selName, objSel + " in " + tmpObj )
selNameRes = "exposed_res_" + randstr
cmd.select(selNameRes, "byres " + selName )
if doShow!=False:
cmd.show_as("spheres", objSel + " and poly")
cmd.color("white", objSel)
cmd.color("red", selName)
cmd.delete(tmpObj)
return exposed
cmd.extend("findSurfaceResidues", findSurfaceResidues)
Another version
from pymol import cmd, stored
def surfaceatoms(molecule="NIL",show=True, verbose=True, cutoff=2.5):
"""
surfaceatoms
finds those residues on the surface of a protein
that have at least 'cutoff' exposed A**2 surface area.
PARAMS
molecule (string)
the object or selection in which to find
exposed residues
DEFAULT: (last molecule in pymol)
cutoff (float)
your cutoff of what is exposed or not.
DEFAULT: 2.5 Ang**2
RETURNS
(list: (chain, resv ) )
A Python list of residue numbers corresponding
to those residues w/more exposure than the cutoff.
"""
if molecule=="NIL":
assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
molecule=cmd.get_names()[-1]
tmpObj="__tmp"
cmd.create(tmpObj, "(%s and polymer) and not resn HOH"%molecule)
if verbose!=False:
print "WARNING: I'm setting dot_solvent. You may not care for this."
cmd.set("dot_solvent")
cmd.get_area(selection=tmpObj, load_b=1)
# threshold on what one considers an "exposed" atom (in A**2):
cmd.remove( tmpObj + " and b < " + str(cutoff) )
stored.tmp_dict = {}
cmd.iterate(tmpObj, "stored.tmp_dict[(chain,resv)]=1")
exposed = stored.tmp_dict.keys()
exposed.sort()
selName = "%s_atoms"%molecule
cmd.select(selName, molecule + " in " + tmpObj )
if verbose!=False:
print "Exposed residues are selected in: " + selName
selNameRes = "%s_resi"%molecule
cmd.select(selNameRes, "byres " + selName )
if show!=False:
cmd.hide("everything", molecule)
cmd.show("cartoon", "%s and not %s and not resn HOH"%(molecule,selNameRes))
cmd.show("sticks", "%s"%selNameRes)
cmd.util.cbaw(selNameRes)
cmd.disable(selNameRes)
#cmd.alter('%s'%(selName),'vdw=0.5') # affects repeated runs
cmd.set('sphere_scale','0.3','%s'%(selName)) # does not affect repeated runs
cmd.show("spheres", "%s"%selName)
cmd.util.cbao(selName)
cmd.disable(selName)
cmd.delete(tmpObj)
print(exposed)
return(exposed)
cmd.extend("surfaceatoms", surfaceatoms)