This is a read-only mirror of pymolwiki.org
Difference between revisions of "Display CCP4 Maps"
| Line 3: | Line 3: | ||
1. Use '''fft''' to create a map in CCP4. (This task can be run from the ''Run | 1. Use '''fft''' to create a map in CCP4. (This task can be run from the ''Run | ||
FFT-Create Map'' option under ''Map & Mask Utilities'') You can create a | FFT-Create Map'' option under ''Map & Mask Utilities'') You can create a | ||
| − | simple map or a 'Fo-Fc map as you wish. CCP4i will create a 2Fo-Fc map by default | + | simple map or a 'Fo-Fc map as you wish. CCP4i will create a 2Fo-Fc map by default. |
To make a Fo-Fc map, set F1=DELFWT and PHIC=PHDELWT in the task window. Select the option to cover 'all | To make a Fo-Fc map, set F1=DELFWT and PHIC=PHDELWT in the task window. Select the option to cover 'all | ||
atoms in PDB file'. For pymol to read the map later, you will need to | atoms in PDB file'. For pymol to read the map later, you will need to | ||
Revision as of 19:51, 21 October 2009
The following steps can be used to read CCP4-format electron density maps into pymol for the generation of publication-quality images.
1. Use fft to create a map in CCP4. (This task can be run from the Run FFT-Create Map option under Map & Mask Utilities) You can create a simple map or a 'Fo-Fc map as you wish. CCP4i will create a 2Fo-Fc map by default. To make a Fo-Fc map, set F1=DELFWT and PHIC=PHDELWT in the task window. Select the option to cover 'all atoms in PDB file'. For pymol to read the map later, you will need to add the file extension .map.ccp4 to the generated map. You may find this map too coarse to make for a pretty diagram. You can generate a nicer (albeit much larger) map by re-running fft with a tighter grid spacing. I usually take the default values generated by FFT and double them. You can change grid spacing under Infrequently Used Options. Alternatively, you can increase the sampling rate in Pymol in step 3 below.
2. Open pymol and read in your PDB file. Create a selection about whatever you want to see the map around. Typically one would display the selection as sticks.
3. Open your map in pymol, e.g. mymap.map.ccp4 (you need the ccp4 extension). An object named mymap.map will be created by pymol. To increase the sampling rate of the map (desirable if you did not do this at the time of map generation) issue the command map_double mymap.map, -1
4. Identify a selection about which to display your map, e.g.
select site, resi 39-50 and resn bct
5. To display the map around your selection (e.g., named site) issue a command similar to this:
isomesh map, mymap.map, 2.0, site, carve=1.6
This command will create a mesh map object named map from the object mymap.map, contoured at 2.0 sigma, around the selection site, within 1.6A of selected atoms. You can change the contour level and carve parameter to suit. Note: the map object will be named in pymol whatever the filename was minus the .ccp4 extension
6. For a publication quality figure the following are suggestions:
color grey50, map # sets map to 50% gray
set mesh_width, 0.5 # makes meshes thinner for ray-tracing
bg_color white #sets background to white
set ray_trace_fog, 0 # turns off raytrace fog--optional
set depth_cue, 0 # turns off depth cueing--optional
set ray_shadows, off # turns off ray-tracing shadows
ray 1024 1024 # this would create a 1024x1024 pixel ray-traced image
png image.png # output final image
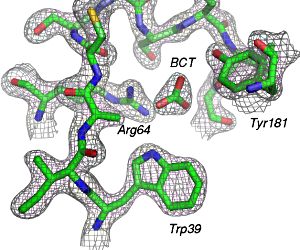
Labels can be added in GIMP or Photoshop. With these directions you should be able to create an image similar to the following one:
User Notes
If the ccp4 maps loaded by PyMOL seem smaller/larger than maps in other programs this might be due to normalization. To stop PyMOL from normalizing a cpp4 map simply unset,
unset normalize_ccp4_maps
before loading your ccp4 map.