This is a read-only mirror of pymolwiki.org
Difference between revisions of "ColorByRMSD"
Jump to navigation
Jump to search
m (Shift-Reload) |
m (→Examples) |
||
| Line 18: | Line 18: | ||
<gallery heights="300px" widths="300px"> | <gallery heights="300px" widths="300px"> | ||
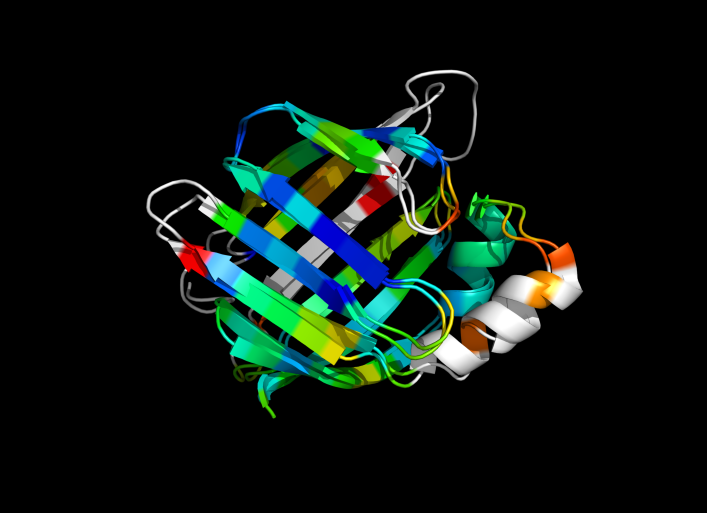
| − | Image:ColorByRMSD_1cbs_1hmt.png|1cbs and 1hmt aligned and colored by RMSD. Dark blue is good alignment, higher deviations are in orange/yellow/red. | + | Image:ColorByRMSD_1cbs_1hmt.png|1cbs and 1hmt aligned and colored by RMSD. Dark blue is good alignment, higher deviations are in orange/yellow/red. Residues not used for alignment are colored white. |
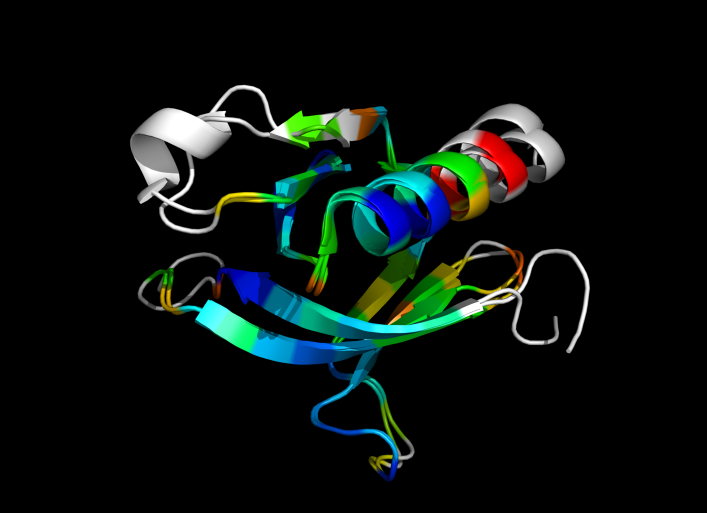
| − | Image:ColorByRMSD_1eaz_1fao.png|1eaz and 1fao aligned and colored by RMSD. Dark blue is good alignment, higher deviations are in orange/yellow/red. | + | Image:ColorByRMSD_1eaz_1fao.png|1eaz and 1fao aligned and colored by RMSD. Dark blue is good alignment, higher deviations are in orange/yellow/red. Residues not used for alignment are colored white. |
</gallery> | </gallery> | ||
Revision as of 20:16, 16 July 2009
THIS PAGE IS UNDERGOING RAPID CHANGES.
PLEASE KEEP REFRESHING YOUR BROWSER's CACHE TO DISPLAY RECENT CHANGES (SHIFT-RELOAD in Firefox).
Introduction
An attempt to perform a coloring of two structures by RMS deviation as calculated by PyMol's internal Rms_Cur command.
Code
This is new code, modified from the work done by Jason and myself.
If you use the option doPretty=T, the residues NOT used for alignment/superposition are now colored white.
Please test and see if it works (somewhat) better. --shiven 19:38, 16 July 2009 (UTC)
Examples
# example #1
colorByRMSD 1cbs, 1hmt, doAlign=True, doPretty=T
# example #2
colorByRMSD 1eaz, 1fao, doAlign=True, doPretty=T
"""
--- ColorByRMSD: RMSD based coloring ---
Authors : Shivender Shandilya; Jason Vertrees
Program : ColorByRMSD
Date : July 2009
Version : 0.0.3 (very alpha!)
Mail : firstname.lastname@umassmed.edu
Keywords: color rms rmsd colorbyrms colorbyrmsd
----------------------------------------------------------------------
Reference:
This email from Warren - http://www.mail-archive.com/pymol-users@lists.sourceforge.net/msg07078.html
Literature:
DeLano, W.L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org
----------------------------------------------------------------------
"""
import pymol
import cmd
from pymol import stored
def strTrue(p):
return p[0].upper() == "T"
# The main function that assigns "cur_rms" as the new b-factor
def rmsUpdateB(objA, alnAri, objB, alnBri):
# don't need the *10 -- PyMOL scales things for us.
for x in range(len(alnAri)):
s1 = objA + " and n. CA and i. " + alnAri[x]
s2 = objB + " and n. CA and i. " + alnBri[x]
rmsd = cmd.rms_cur(s1, s2, matchmaker=4)
cmd.alter( s1, "b = " + str(rmsd))
cmd.alter( s2, "b = " + str(rmsd))
cmd.sort(objA); cmd.sort(objB)
def colorByRMSD(objSel1, objSel2, doAlign="True", doPretty=None):
"""
colorByRMSD -- align two structures and show the structural deviations in
color to more easily see variable regions.
PARAMS
objSel1 (valid PyMOL object or selection)
The first object to align.
objSel2 (valid PyMOL object or selection)
The second object to align
doAlign (boolean, either True or False)
Should this script align your proteins or just leave them where they
are? If doAlign=True then your original proteins are aligned. If
doAlign=False, then they are not. Regardless, the b-factors are
changed.
DEFAULT: True
doPretty (boolean, either True or False)
If doPretty=True then a simple representation is created to high-
light the differences. If False, then no change is done to the
structure.
DEFAULT: None
RETURNS
None.
SIDE-EFFECTS
Modified the b-factor columns in your original proteins.
"""
# create backup copies; names starting with _ (underscores) are
# hidden by PyMOL
tObj1, tObj2, aln = "__tempObj1", "__tempObj2", "__aln"
if strTrue(doAlign):
# perform the alignment
cmd.create( tObj1, objSel1 )
cmd.create( tObj2, objSel2 )
# cmd.align( tObj1, tObj2, object=aln )
cmd.super( tObj1, tObj2, object=aln )
# bug -- every other call undoes this...
cmd.matrix_copy(tObj1, objSel1)
else:
# perform the alignment
cmd.create( tObj1, objSel1 )
cmd.create( tObj2, objSel2 )
# cmd.align( tObj1, tObj2, object=aln )
cmd.super( tObj1, tObj2, object=aln )
# cmd.alter( tObj1 + " or " + tObj2, "b=-10")
# Now modify the B-factor columns of the original objects,
# so as to later identify the residues NOT used for alignment
cmd.alter( objSel1 + " or " + objSel2, "b=-10")
cmd.alter( tObj1 + " or " + tObj2, "chain='A'")
cmd.alter( tObj1 + " or " + tObj2, "segi='A'")
# update PyMOL;
# one of these should do the trick
cmd.refresh(); cmd.rebuild(); cmd.sort(tObj1); cmd.sort(tObj2)
# Get the residue identifiers from the aln object
stored.alnAres, stored.alnBres = [], []
cmd.iterate(tObj1 + " and n. CA and " + aln, "stored.alnAres.append(resi)")
cmd.iterate(tObj2 + " and n. CA and " + aln, "stored.alnBres.append(resi)")
#print "Length of alnAres using 'super': "+str(len(stored.alnAres))
#print "Length of alnBres using 'super': "+str(len(stored.alnBres))
# reset the b-factors for each object
rmsUpdateB(tObj1,stored.alnAres,tObj2,stored.alnBres)
# Store the NEW b-factors
stored.alnAnb, stored.alnBnb = [], []
cmd.iterate(tObj1 + " and n. CA and " + aln, "stored.alnAnb.append(b)" )
cmd.iterate(tObj2 + " and n. CA and " + aln, "stored.alnBnb.append(b)" )
# Get rid of all intermediate objects etc.; clean up
cmd.delete(tObj1)
cmd.delete(tObj2)
cmd.delete(aln)
# Assign the just stored NEW b-factors to the original object
for x in range(len(stored.alnAres)):
cmd.alter(objSel1 + " and n. CA and i. " + str(stored.alnAres[x]), "b = " + str(stored.alnAnb[x]))
for x in range(len(stored.alnBres)):
cmd.alter(objSel2 + " and n. CA and i. " + str(stored.alnBres[x]), "b = " + str(stored.alnBnb[x]))
cmd.rebuild(); cmd.refresh(); cmd.sort(objSel1); cmd.sort(objSel2)
if doPretty!=None:
# Showcase what we did
cmd.orient()
cmd.hide("all")
cmd.show_as("cartoon", objSel1 + " or " + objSel2)
# Select the residues not used for alignment; they still have their B-factors as "-10"
cmd.select("notUsedForAln", "b < 0")
cmd.disable("notUsedForAln")
# White-wash the residues not used for alignment
cmd.color("white", "notUsedForAln")
# Color the residues used for alignment according to their B-factors (RMSD values)
cmd.spectrum("b", 'rainbow', "((" + objSel1 + " and n. CA) or (n. CA and " + objSel2 +" )) and not notUsedForAln")
print "\nColorByRMSD completed successfully. Objects are now colored by C-alpha RMS deviation.\n"
cmd.extend("colorByRMSD", colorByRMSD)