This is a read-only mirror of pymolwiki.org
Difference between revisions of "ColorByDisplacement"
(Created page with '== Acknowledgement == This script is based on the scaffold from ColorByRMSD. Peace love and harmomy goes to Shivender Shandilya and Jason Vertrees. == Introduction == This scrip…') |
|||
| Line 7: | Line 7: | ||
== Code == | == Code == | ||
Please use this script with the option '''doColor=T''' to print the coloring. Do keep in mind, all original B-factors values are overwritten! | Please use this script with the option '''doColor=T''' to print the coloring. Do keep in mind, all original B-factors values are overwritten! | ||
| − | There exist two versions. ColorByDisplacementCA is quick and is between CA atoms. Ideal for helices representation. ColorByDisplacementAll is between All atoms in residues and is quite slow => 3-5 mins for a run. Ideal for sticks representation. | + | |
| + | There exist two versions. <br> | ||
| + | ColorByDisplacementCA is quick and is between CA atoms. Ideal for helices representation. <br> | ||
| + | ColorByDisplacementAll is between All atoms in residues and is quite slow => 3-5 mins for a run. Ideal for sticks representation. | ||
Revision as of 20:20, 4 January 2011
Acknowledgement
This script is based on the scaffold from ColorByRMSD. Peace love and harmomy goes to Shivender Shandilya and Jason Vertrees.
Introduction
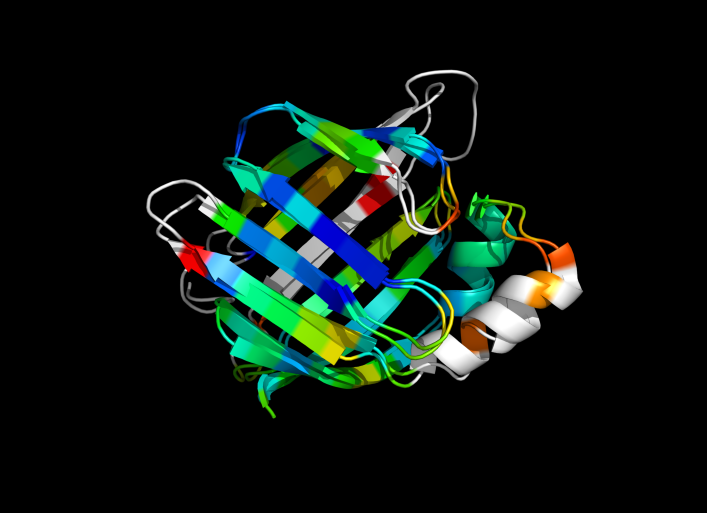
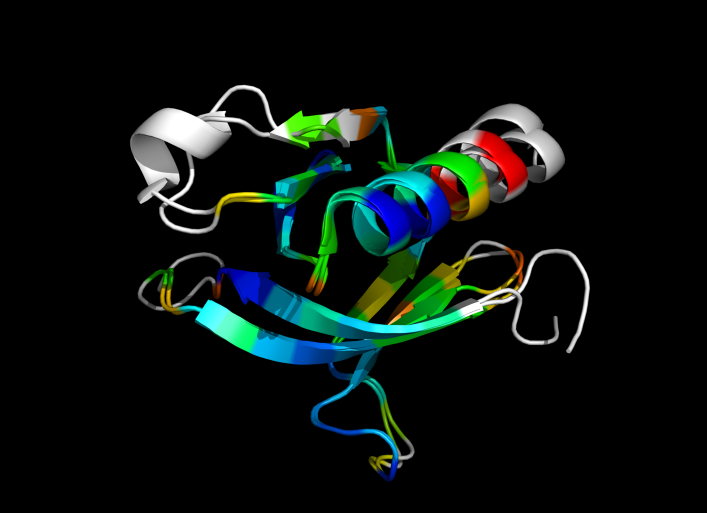
This script allows you to color two structures by distance displacement between a Open and Closed form of a protein, as calculated by PyMol's internal distance command. The pairwise, C-alpha or all-atom. . The RMSD values are stored as B-factors of these residues, which are colored by a rainbow color spectrum, with blue specifying the minimum pairwise RMSD and red indicating the maximum. With uncommenting/commenting it is possible to make Residues NOT used by Super for superposition, and hence for distance displacement calculation, colored white.
Code
Please use this script with the option doColor=T to print the coloring. Do keep in mind, all original B-factors values are overwritten!
There exist two versions.
ColorByDisplacementCA is quick and is between CA atoms. Ideal for helices representation.
ColorByDisplacementAll is between All atoms in residues and is quite slow => 3-5 mins for a run. Ideal for sticks representation.
Examples
# example #1
ColorByDisplacementCA O5NT, C5NT, doColor=T, doAlign=T, super1=resi 26-355, super2=resi 26-355
ColorByDisplacementAll O5NT, C5NT, doColor=T, doAlign=T, super1=resi 26-355, super2=resi 26-355
"""
--- ColorByRMSD: RMSD based coloring ---
Authors : Shivender Shandilya; Jason Vertrees
Program : ColorByRMSD
Date : July 2009
Version : 0.1.1
Mail : firstname.lastname@umassmed.edu
Keywords: color rms rmsd colorbyrms colorbyrmsd
----------------------------------------------------------------------
Reference:
This email from Warren - http://www.mail-archive.com/pymol-users@lists.sourceforge.net/msg07078.html
Literature:
DeLano, W.L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org
----------------------------------------------------------------------
"""
import pymol
import cmd
from pymol import stored
def strTrue(p):
return p[0].upper() == "T"
# The main function that assigns current RMSD as the new B-factor
def rmsUpdateB(objA, alnAri, objB, alnBri):
for x in range(len(alnAri)):
s1 = objA + " and n. CA and i. " + alnAri[x]
s2 = objB + " and n. CA and i. " + alnBri[x]
rmsd = cmd.rms_cur(s1, s2, matchmaker=4)
cmd.alter( s1, "b = " + str(rmsd))
cmd.alter( s2, "b = " + str(rmsd))
cmd.sort(objA); cmd.sort(objB)
def colorByRMSD(objSel1, objSel2, doAlign="True", doPretty=None):
"""
colorByRMSD -- align two structures and show the structural deviations
in color to more easily see variable regions.
PARAMS
objSel1 (valid PyMOL object or selection)
The first object to align.
objSel2 (valid PyMOL object or selection)
The second object to align
doAlign (boolean, either True or False)
Should this script align your proteins or just leave them as is?
If doAlign=True then your original proteins are aligned.
If False, then they are not. Regardless, the B-factors are changed.
DEFAULT: True
doPretty (boolean, either True or False)
If doPretty=True then a simple representation is created to
highlight the differences. If False, then no changes are made.
DEFAULT: False
RETURNS
None.
SIDE-EFFECTS
Modifies the B-factor columns in your original structures.
"""
# First create backup copies; names starting with __ (underscores) are
# normally hidden by PyMOL
tObj1, tObj2, aln = "__tempObj1", "__tempObj2", "__aln"
if strTrue(doAlign):
# perform the alignment
cmd.create( tObj1, objSel1 )
cmd.create( tObj2, objSel2 )
cmd.super( tObj1, tObj2, object=aln )
cmd.matrix_copy(tObj1, objSel1)
else:
# perform the alignment
cmd.create( tObj1, objSel1 )
cmd.create( tObj2, objSel2 )
cmd.super( tObj1, tObj2, object=aln )
# Modify the B-factor columns of the original objects,
# in order to identify the residues NOT used for alignment, later on
cmd.alter( objSel1 + " or " + objSel2, "b=-10")
cmd.alter( tObj1 + " or " + tObj2, "chain='A'")
cmd.alter( tObj1 + " or " + tObj2, "segi='A'")
# Update pymol internal representations; one of these should do the trick
cmd.refresh(); cmd.rebuild(); cmd.sort(tObj1); cmd.sort(tObj2)
# Create lists for storage
stored.alnAres, stored.alnBres = [], []
# Get the residue identifiers from the alignment object "aln"
cmd.iterate(tObj1 + " and n. CA and " + aln, "stored.alnAres.append(resi)")
cmd.iterate(tObj2 + " and n. CA and " + aln, "stored.alnBres.append(resi)")
# Change the B-factors for EACH object
rmsUpdateB(tObj1,stored.alnAres,tObj2,stored.alnBres)
# Store the NEW B-factors
stored.alnAnb, stored.alnBnb = [], []
cmd.iterate(tObj1 + " and n. CA and " + aln, "stored.alnAnb.append(b)" )
cmd.iterate(tObj2 + " and n. CA and " + aln, "stored.alnBnb.append(b)" )
# Get rid of all intermediate objects and clean up
cmd.delete(tObj1)
cmd.delete(tObj2)
cmd.delete(aln)
# Assign the just stored NEW B-factors to the original objects
for x in range(len(stored.alnAres)):
cmd.alter(objSel1 + " and n. CA and i. " + str(stored.alnAres[x]), "b = " + str(stored.alnAnb[x]))
for x in range(len(stored.alnBres)):
cmd.alter(objSel2 + " and n. CA and i. " + str(stored.alnBres[x]), "b = " + str(stored.alnBnb[x]))
cmd.rebuild(); cmd.refresh(); cmd.sort(objSel1); cmd.sort(objSel2)
# Provide some useful information
stored.allRMSDval = []
stored.allRMSDval = stored.alnAnb + stored.alnBnb
print "\nColorByRMSD completed successfully."
print "The MINIMUM RMSD value is: "+str(min(stored.allRMSDval))
print "The MAXIMUM RMSD value is: "+str(max(stored.allRMSDval))
if doPretty!=None:
# Showcase what we did
cmd.orient()
cmd.hide("all")
cmd.show_as("cartoon", objSel1 + " or " + objSel2)
# Select the residues not used for alignment; they still have their B-factors as "-10"
cmd.select("notUsedForAln", "b < 0")
# White-wash the residues not used for alignment
cmd.color("white", "notUsedForAln")
# Color the residues used for alignment according to their B-factors (RMSD values)
cmd.spectrum("b", 'rainbow', "((" + objSel1 + " and n. CA) or (n. CA and " + objSel2 +" )) and not notUsedForAln")
# Delete the selection of atoms not used for alignment
# If you would like to keep this selection intact,
# just comment "cmd.delete" line and
# uncomment the "cmd.disable" line below.
cmd.delete("notUsedForAln")
# cmd.disable("notUsedForAln")
print "\nObjects are now colored by C-alpha RMS deviation."
print "All residues with RMSD values greater than the maximum are colored white..."
cmd.extend("colorByRMSD", colorByRMSD)