This is a read-only mirror of pymolwiki.org
ShowLigandWaters
Revision as of 08:56, 2 September 2013 by GianlucaTomasello (talk | contribs)
| Type | Python Script |
|---|---|
| Download | |
| Author(s) | Gianluca Tomasello |
| License | CC BY-NC-SA |
This is a test page########
Introduction
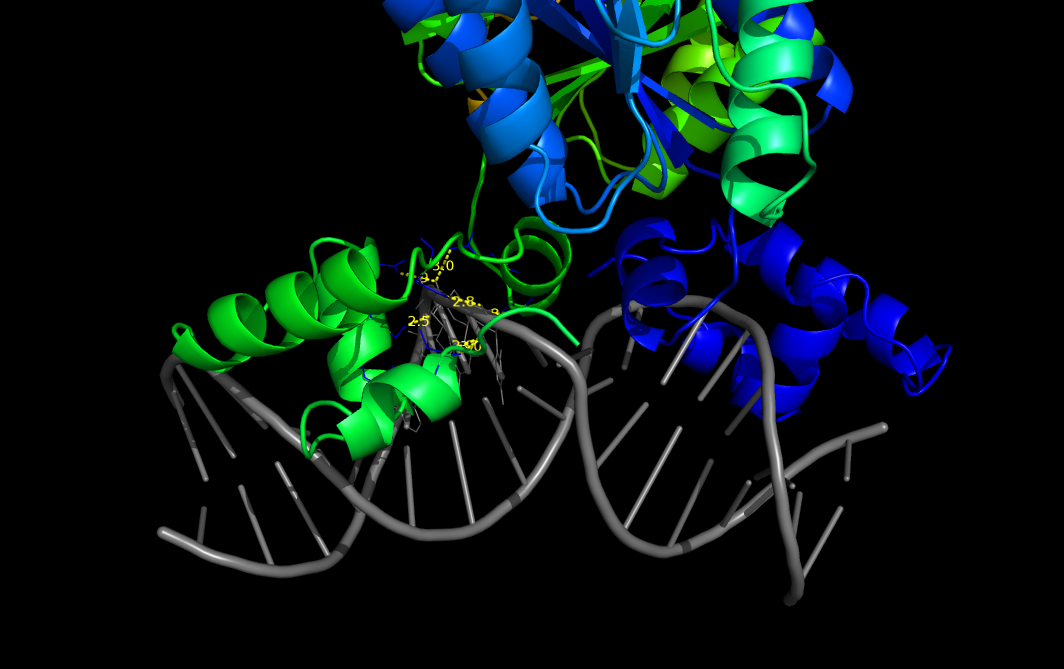
This script detects waters molecules within a specified distance from the ligand. Water molecules are shown. Distance between water molecules and O or N atoms of ligand are shown and is maked an output file containing a list of distance between waters and ligand atoms and the number of interactions
Usage
waters [ligand name, distance]
Required Arguments
- ligand name = the ligand residue name
- distance = max distance in Angstroms
Examples
example #1
PyMOL>pairwise_dist 1efa and chain D, 1efa and chain B, 3, output=S, show=Y
1efa/D/DC/13/OP1 to 1efa/B/TYR/47/OH: 2.765
1efa/D/DC/13/OP2 to 1efa/B/LEU/6/N: 2.983
1efa/D/DC/13/OP2 to 1efa/B/LEU/6/CB: 2.928
1efa/D/DT/14/O4' to 1efa/B/ALA/57/CB: 2.827
1efa/D/DT/14/OP1 to 1efa/B/ASN/25/OD1: 2.858
1efa/D/DT/14/OP1 to 1efa/B/GLN/54/NE2: 2.996
1efa/D/DT/14/OP2 to 1efa/B/SER/21/OG: 2.517
1efa/D/DC/15/N4 to 1efa/B/GLN/18/NE2: 2.723
1efa/D/DA/16/N6 to 1efa/B/GLN/18/NE2: 2.931
Number of distances calculated: 9
The Code
Copy the following text and save it as pairwisedistances.py
from pymol import cmd, stored, math
def pairwise_dist(sel1, sel2, max_dist, output="N", sidechain="N", show="N"):
"""
usage: pairwise_dist sel1, sel2, max_dist, [output=S/P/N, [sidechain=N/Y, [show=Y/N]]]
sel1 and sel2 can be any to pre-existing or newly defined selections
max_dist: maximum distance in Angstrom between atoms in the two selections
--optional settings:
output: accepts Screen/Print/None (default N)
sidechain: limits (Y) results to sidechain atoms (default N)
show: shows (Y) individual distances in pymol menu (default=N)
"""
print ""
cmd.delete ("dist*")
extra=""
if sidechain=="Y": extra=" and not name c+o+n"
#builds models
m1=cmd.get_model(sel2+" around "+str(max_dist)+" and "+sel1+extra)
m1o=cmd.get_object_list(sel1)
m2=cmd.get_model(sel1+" around "+str(max_dist)+" and "+sel2+extra)
m2o=cmd.get_object_list(sel2)
#defines selections
cmd.select("__tsel1a", sel1+" around "+str(max_dist)+" and "+sel2+extra)
cmd.select("__tsel1", "__tsel1a and "+sel2+extra)
cmd.select("__tsel2a", sel2+" around "+str(max_dist)+" and "+sel1+extra)
cmd.select("__tsel2", "__tsel2a and "+sel1+extra)
cmd.select("IntAtoms_"+max_dist, "__tsel1 or __tsel2")
cmd.select("IntRes_"+max_dist, "byres IntAtoms_"+max_dist)
#controlers-1
if len(m1o)==0:
print "warning, '"+sel1+extra+"' does not contain any atoms."
return
if len(m2o)==0:
print "warning, '"+sel2+extra+"' does not contain any atoms."
return
#measures distances
s=""
counter=0
for c1 in range(len(m1.atom)):
for c2 in range(len(m2.atom)):
distance=math.sqrt(sum(map(lambda f: (f[0]-f[1])**2, zip(m1.atom[c1].coord,m2.atom[c2].coord))))
if distance<float(max_dist):
s+="%s/%s/%s/%s/%s to %s/%s/%s/%s/%s: %.3f\n" % (m1o[0],m1.atom[c1].chain,m1.atom[c1].resn,m1.atom[c1].resi,m1.atom[c1].name,m2o[0],m2.atom[c2].chain,m2.atom[c2].resn,m2.atom[c2].resi,m2.atom[c2].name, distance)
counter+=1
if show=="Y": cmd.distance (m1o[0]+" and "+m1.atom[c1].chain+"/"+m1.atom[c1].resi+"/"+m1.atom[c1].name, m2o[0]+" and "+m2.atom[c2].chain+"/"+m2.atom[c2].resi+"/"+m2.atom[c2].name)
#controler-2
if counter==0:
print "warning, no distances were measured! Check your selections/max_dist value"
return
#outputs
if output=="S": print s
if output=="P":
f=open('IntAtoms_'+max_dist+'.txt','w')
f.write("Number of distances calculated: %s\n" % (counter))
f.write(s)
f.close()
print "Results saved in IntAtoms_%s.txt" % max_dist
print "Number of distances calculated: %s" % (counter)
cmd.hide("lines", "IntRes_*")
if show=="Y": cmd.show("lines","IntRes_"+max_dist)
cmd.deselect()
cmd.extend("pairwise_dist", pairwise_dist)