This is a read-only mirror of pymolwiki.org
Difference between revisions of "Annotation wizard"
Jump to navigation
Jump to search
m (4 revisions) |
(No difference)
|
Latest revision as of 01:06, 28 March 2014
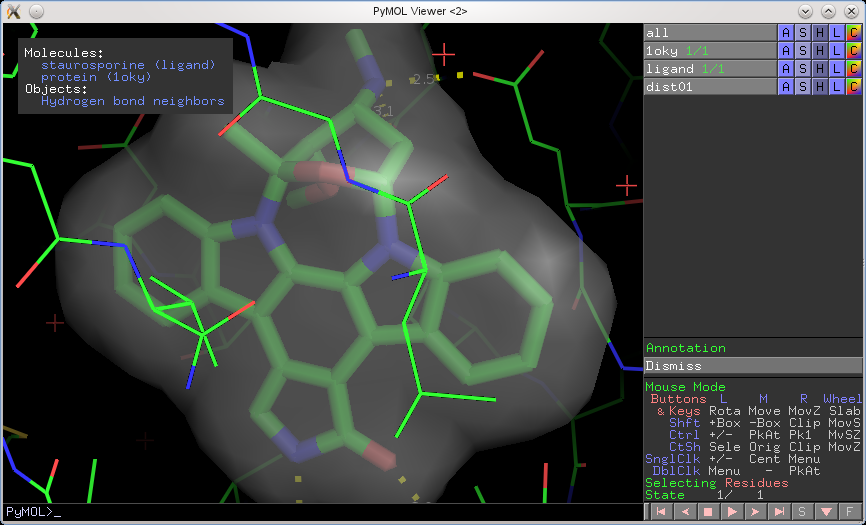
Here's an example of using the annotation wizard (see top-left corner):
import pymol
# fetch a protein and setup the view
cmd.fetch("1oky", async=0)
cmd.extract("ligand", "resn STU")
cmd.show_as("sticks", "resn STU")
cmd.show("surface", "ligand")
cmd.flag("ignore", "not rep surface")
cmd.set("surface_color", "grey")
cmd.set("transparency", 0.3)
cmd.distance("(ligand)", "(poly)", quiet=1, mode=2, label=1)
# turn on the annotation wizard + prompt
cmd.wizard("annotation")
cmd.set("wizard_prompt_mode", 1)
pymol.session.annotation = {}
state_dict = {1: ['\\999Molecules:',' \\459staurosporine (ligand)',' \\459protein (1oky)','\\999Objects:',' \\459Hydrogen bond neighbors',]}
pymol.session.annotation["1oky"] = state_dict
The following source shows you how to use the annotation wizard for multiple objects.
import pymol
# fetch a protein and setup the view
cmd.fetch("1oky", async=0)
cmd.extract("ligand", "resn STU")
cmd.show_as("sticks", "resn STU")
cmd.show("surface", "ligand")
cmd.flag("ignore", "not rep surface")
cmd.set("surface_color", "grey")
cmd.set("transparency", 0.3)
cmd.distance("(ligand)", "(poly)", quiet=1, mode=2, label=1)
# turn on the annotation wizard + prompt
cmd.wizard("annotation")
cmd.set("wizard_prompt_mode", 1)
pymol.session.annotation = {}
# using the annotation wizard for multiple objects:
prot_dict = { 1: ['\\999Protein:',' \\459protein (1oky)',]}
lig_dict = { 1: ['\\999Ligand:',' \\459staurosporine (ligand)',]}
dist_dict = { 1: ['\\999Objects: ',' \\459Hydrogen bond neighbors',]}
pymol.session.annotation["1oky"] = prot_dict
pymol.session.annotation["ligand"] = lig_dict
pymol.session.annotation["dist01"] = dist_dict