This is a read-only mirror of pymolwiki.org
Difference between revisions of "ConnectedCloud"
Jump to navigation
Jump to search
m (7 revisions) |
(No difference)
|
Latest revision as of 01:21, 28 March 2014
Overview
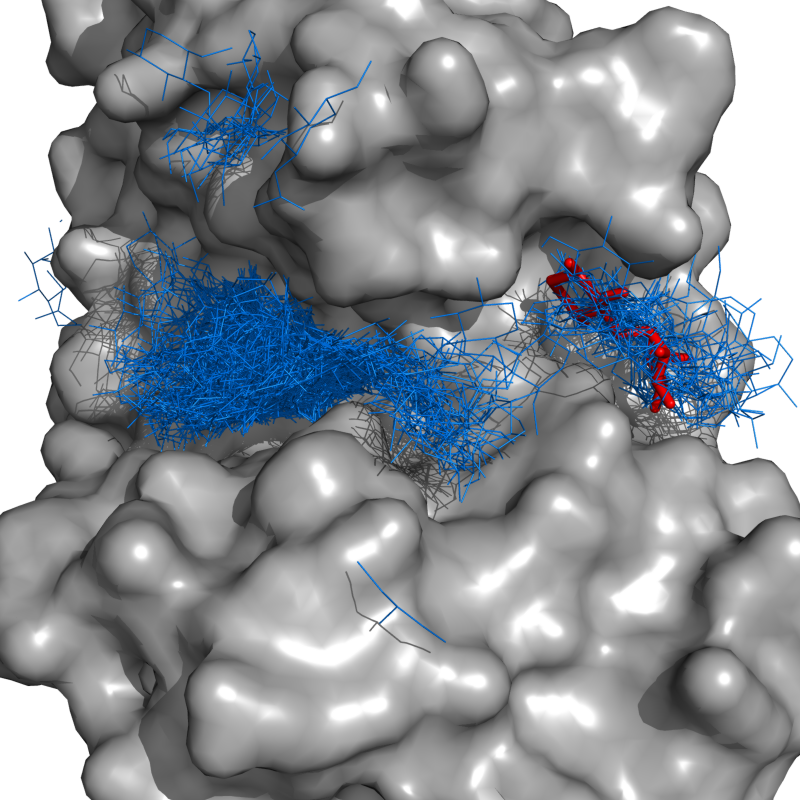
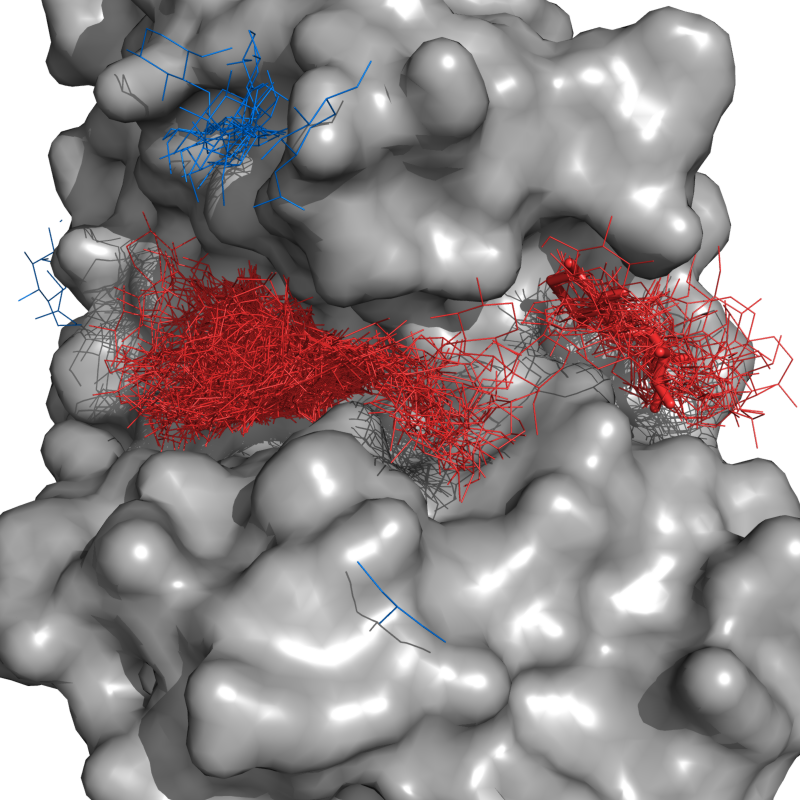
ConnectedCloud grows a cloud of objects that are "connected" through single-linkage clustering. Two objects are "connected" if they are within the user-defined cutoff, radius. It starts with a selection, and grows until no more objects are withing radius Angstroms of the cloud.
Let's say we want to find the cloud of docking guesses (blue lines) that are connected to the lowest energy docking pose (red sticks). I call,
connectedCloud lig1, lig*, radius=1, doPrint=True
and get
Syntax
connectedCloud origSel, subSel, [radius=1, [doPrint=True]]
- origSel -- the original selection to grow from
- subSel -- only consider these objects when growing
- radius -- defines "connectedness". Anything <= radius Angstroms from the growing cloud and in subSel is considered
- doPrint -- print some misc. info
Examples
# Find all objects in the set "lig*" (all ligands from a docking run, for example) that are
# in the connected cloud of "connectedness" set to 1 Ang. Print the number of atoms in the
# selection and the surface area of the selection (if it were an object).
connectedCloud lig427, lig*, radius=1, doPrint=True
# select everything within 0.85 of lig2467 and don't print anything
connectedCloud lig2467, *, radius=0.85, doPrint=False
The Code
# -*- coding: utf-8 -*-
def connectedCloud( origSel, subSel, radius=1, doPrint=True ):
"""
connectedCloud -- find all atoms, by object in subSel connected to
origSel by radius Ang. In other words, start from origSel
and find all objects <=radius Ang. from it & add that to the
result. Once nothing more is added, you have your cloud of
points.
origSel,
the selection around which we start expanding.
subSel,
the subset of objects to consider
radius,
the maximum radius in which to consider; anything >radius
Ang. from any objet in the current selection is ignored (unless
it is added later by some other path).
doPrint
print some values before returning the selection name
Author:
Jason Vertrees, 2009.
"""
cName = "__tempCloud__"
cmd.select(cName, origSel)
# we keep track of cloud size based on the number of atoms in the selection; oldCount
# is the previous iteration and curCount is after we add everything for this iteration.
oldCount=0
curCount=cmd.count_atoms( cName )
# flag to stop at 1000 iterations. if something goes wrong we don't want to hijack the computer.
flag=0
# while something was added in this iteration
while oldCount != curCount:
flag += 1
# grow current cloud
cmd.select( cName, "bo. " + subSel + " within " + radius + " of " + cName )
# update atom counts in the cloud
oldCount=curCount
curCount=cmd.count_atoms(cName)
if ( flag > 1000 ):
print "Warning: I gave up. I grew the cloud 1000 times and it kept growing."
print "Warning: If this is valid (weird), then increases the 'flag' in the source"
print "Warning: code to something >1000."
break
# create the object for getting its surface area
cmd.create("connectedCloud", cName)
# print cloud info
if doPrint:
print "Number of atoms in cloud:", str(cmd.count_atoms(cName))
print "Surface area of cloud:", str(cmd.get_area("connectedCloud"))
# delete our work
cmd.delete("connectedCloud")
# rename and return
cmd.set_name(cName, "connectedCloud")
return "connectedCloud"
cmd.extend("connectedCloud", connectedCloud)