This is a read-only mirror of pymolwiki.org
Difference between revisions of "Optimize"
m (5 revisions) |
(change license) |
||
| Line 3: | Line 3: | ||
|filename = plugins/optimize.py | |filename = plugins/optimize.py | ||
|author = [[User:OsvaldoMartin|Osvaldo Martin]] | |author = [[User:OsvaldoMartin|Osvaldo Martin]] | ||
| − | |license = | + | |license = MIT |
}} | }} | ||
Revision as of 08:15, 14 June 2017
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/optimize.py |
| Author(s) | Osvaldo Martin |
| License | MIT |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
Optimize provides a PyMOL graphical interface to some of the molecular mechanics features available in openbabel, allowing the user to optimize (minimize) the energy of any molecule uploaded on PyMOL.
Installation
The plugin can be downloaded through the project Pymol-script-repo.
Optimize needs OpenBabel (and OpenBabel Python bindings) to be installed on your computer (see instructions here).
Note to Windows users: If you are using a 64-bit Windows, please install OpenBabel 64-bit from here
Usage
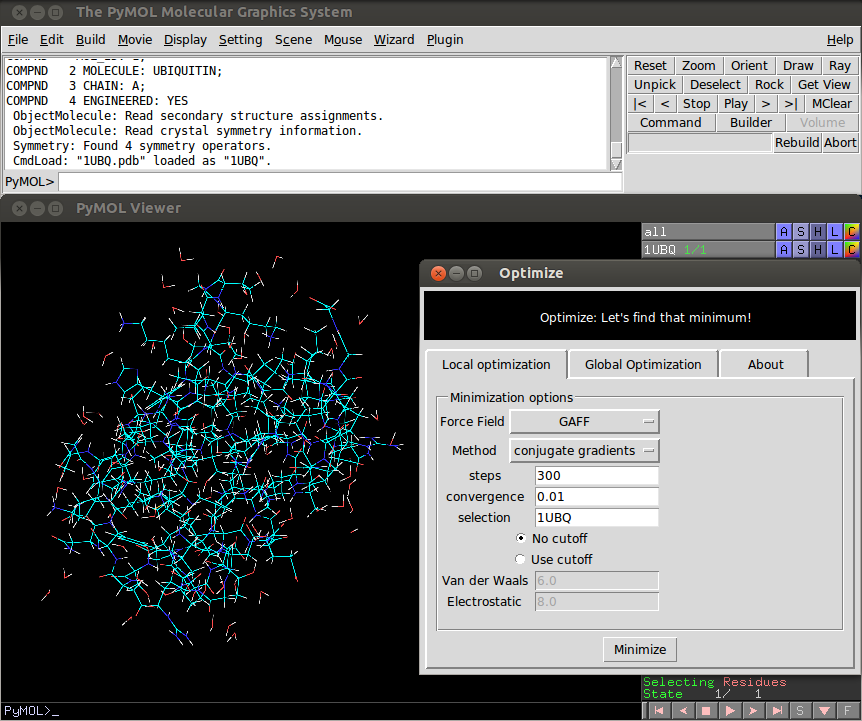
The plugin can be accessed using a graphic user interface (see figure 0) or from the PyMOL`s terminal. There are 5 types of optimization routines available now, 2 local and 3 global optimization routines.
Local optimization
Local optimization can be done using the minimize command.
Usage
minimize [selection [, forcefield [, method [, nsteps [, conv [, cutoff [, cut_vdw [, cut_elec]]]]]]]]
- selection: The name of the object to be minimized. From the command line, the default value is 'all'. When using the GUI, the default value is the first uploaded object.
- forcefield: The forcefield used to compute the Internal Energy. Available options are GAFF, MMFF94s (default), MMFF94, UFF and Ghemical.
- method: The method used to find the local minimum. Available options are "conjugate gradients" (default) and "steepest descent".
- nsteps: Number of iteration steps during the minimization (default = 500).
- conv: Criteria used to judge minimization convergence (default = 0.0001).
- cutoff: Control if cut-off are used or not to compute non-bonded interactions, possible values are True or False (default).
- cut_vdw: If cutoff is True, then this parameter sets the distance (in Angstroms) beyond which two atoms do not interact through Van der Waals forces (default = 6.0).
- cut_elec: If cutoff is True, then this parameter sets the distance (in Angstroms) beyond which two atoms do not interact through electrostatic forces (default = 8.0).
PyMOL API
cmd.minimize(string selection="all", string forcefield="MMFF94s", string method="conjugate gradients",
int nsteps=500, float conv=0.0001, bool cutoff=False, float cut_vdw=6.0, float cut_elec=8.0)
Global optimization
Global optimization can be done using the conf_search command from the PyMOL`s terminal:
conf_search [selection string [, forcefield string [, method string [, nsteps int [, conformers int [, lowest_confor int]]]]]]
Where:
- selection: The name of teh object that is going to be minimized. The default value is 'all'. Using the GUI the default value is the first uploaded object.
- forcefield: Choose the forcefield used to compute the Internal Energy, options available are GAFF, MMFF94s, MMFF94, UFF and Ghemical.
- method: Choose the method used to find the global minimum. The methods available are:
Systematic: Systematically iterate through all possible conformers according to Open Babel’s torsion library. This approach guarantee to find the local minimum (according to the forcefield in use). This approach scales to the power of N, where N is the number of rotatable bonds, hence it is only applicable to molecules with very few rotatable bonds.
Random: Conformations are generated by randomly choosing from the allowed torsion angles.
Weighted: This method uses an iterative procedure to find a global minimum. As with the Random method, it randomly choses from the allowed torsion angles but the choice is re-weighted based on the energy of the generated conformer. For molecules with to many rotatable bonds, that are not suitable for for the systematic, this method is generally the best option.
- nsteps: Number of iteration steps during the minimization.
- conformers: Total number of conformers to be analysed. This option is not available when using the systematic method because all possible conformers are analysed.
- lowest_conf: This options sets how many of the low-energy conformers are retrieved as the result of a conformational search. Conformers are ordered from low to high energy. This option is not available when using the systematic method because this method return only the lowest energy conformer.
In general, it is a good idea to minimize the initial conformation before doing a conformational search.
Change log
- 2013-10-06 (Version 0.1)
- First version. (More features coming soon!)
- 2013-10-24 (Version 0.2)
- now openbabel add the hydrogen and not PyMOL.
- 2014-01-27 (Version 0.6)
- now it is possible to perform global optimization.
- 2014-08-22 (Version 0.8)
- Bug fixed. In version 0.6 the 'selection' and 'steps' parameters for the local minimization were incorrectly taken from the gobal optimization tab
Citation
If you find optimize useful please consider citing this work Noel M. O'Boyle , Michael Banck , Craig A. James , Chris Morley , Tim Vandermeersch and Geoffrey R. Hutchison. "Open Babel: An open chemical toolbox." Journal of Cheminformatics 2011, 3:33. http://www.jcheminf.com/content/3/1/33