|
|
| (6 intermediate revisions by one other user not shown) |
| Line 1: |
Line 1: |
| − | == Author == | + | {{Infobox script-repo |
| − | This pymol script is made by Troels Emtekær Linnet
| + | |type = script |
| | + | |filename = rotkit.py |
| | + | |author = [[User:Tlinnet|Troels E. Linnet]] |
| | + | |license = BSD |
| | + | }} |
| | | | |
| | == Introduction == | | == Introduction == |
| Line 53: |
Line 57: |
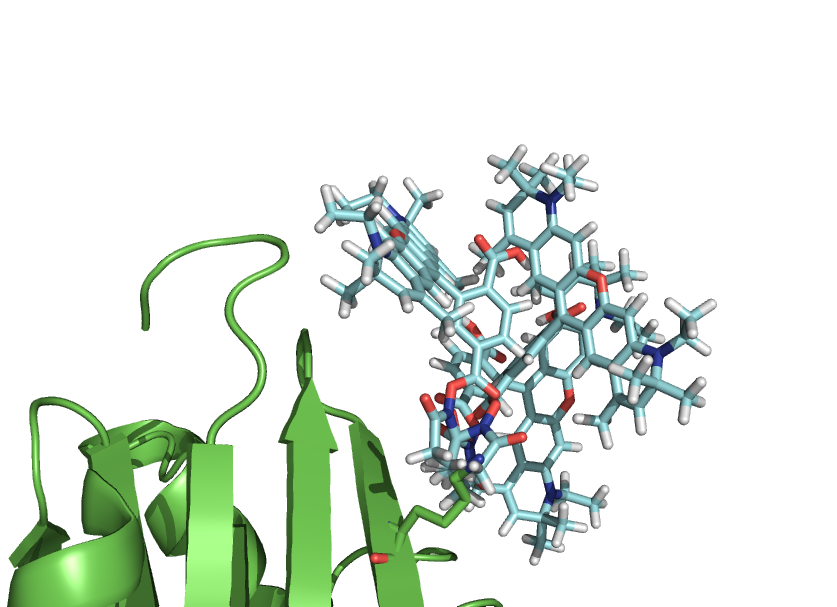
| | == Example of use == | | == Example of use == |
| | === Example 1 - Make a rotation of domain === | | === Example 1 - Make a rotation of domain === |
| − | <syntaxhighlight lang="python">
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_1.pml}} |
| − | reinitialize
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_1.pml" highlight="python" /> |
| − | import rotkit
| |
| − |
| |
| − | fetch 1HP1, async=0
| |
| − | show_as cartoon, 1HP1
| |
| − | show_as sticks, 1HP1 and resn ATP
| |
| − |
| |
| − | ###################### Make rotation axis #################
| |
| − | pseudoatom axisA, vdw=1.0
| |
| − | pseudoatom axisB, vdw=1.0
| |
| − | rotkit.toline("/1HP1//A/477/C","/1HP1//A/423/CG1","axisA","axisA",20)
| |
| − | rotkit.toline("/1HP1//A/423/CG1","/1HP1//A/477/C","axisB","axisB",5)
| |
| − | show spheres, axisA or axisB
| |
| − | label axisA, "axisA"
| |
| − | label axisB, "axisB"
| |
| − | dist rotaxis, axisA, axisB
| |
| − | color green, rotaxis
| |
| − | set dash_width, 5
| |
| − | set dash_gap, 0
| |
| − | hide label, rotaxis
| |
| − |
| |
| − | ## Create rotate states of 1HP1
| |
| − | create 1HP1_rot, 1HP1, 1, 1
| |
| − | python | |
| − | ang_incr = 1
| |
| − | anglerange = range(2,98,ang_incr)
| |
| − | nrstates = len(anglerange)+1
| |
| − | states = 1
| |
| − | for angle in anglerange:
| |
| − | states += 1
| |
| − | rot_1HP1 = "1HP1_rot_%s"%angle
| |
| − | cmd.create(rot_1HP1,"(1HP1 and resi 363-550) or (1HP1 and resn ATP)")
| |
| − | rotkit.rotateline("axisA","axisB",-(angle-1),rot_1HP1)
| |
| − | cmd.create("1HP1_rot",rot_1HP1,1,states)
| |
| − | cmd.create("1HP1_rot",rot_1HP1,1,2*nrstates-states)
| |
| − | cmd.delete(rot_1HP1)
| |
| − | python end
| |
| − | hide cartoon, (1HP1 and resi 363-550)
| |
| − | hide sticks, (1HP1 and resn ATP)
| |
| − | mplay
| |
| − | </syntaxhighlight>
| |
| | | | |
| | === Example 2 - Simulate dye freedom === | | === Example 2 - Simulate dye freedom === |
| − | <syntaxhighlight lang="python">
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_2.pml}} |
| − | reinitialize
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_2.pml" highlight="python" /> |
| − | import rotkit
| |
| − |
| |
| − | fetch 1HP1, async=0
| |
| − | show_as cartoon, 1HP1
| |
| − | show_as sticks, 1HP1 and resn ATP
| |
| − | set auto_zoom, off
| |
| − |
| |
| − | ###################### Make rotation axis #################
| |
| − | pseudoatom axisA, vdw=1.0
| |
| − | pseudoatom axisB, vdw=1.0
| |
| − | rotkit.toline("/1HP1//A/477/C","/1HP1//A/423/CG1","axisA","axisA",20)
| |
| − | rotkit.toline("/1HP1//A/423/CG1","/1HP1//A/477/C","axisB","axisB",5)
| |
| − | show spheres, axisA or axisB
| |
| − | label axisA, "axisA"
| |
| − | label axisB, "axisB"
| |
| − | dist rotaxis, axisA, axisB
| |
| − | color green, rotaxis
| |
| − | set dash_width, 5
| |
| − | set dash_gap, 0
| |
| − | hide label, rotaxis
| |
| − |
| |
| − | ####################### Create rotate states of dye atoms ###################
| |
| − | ##### First mutate, the mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
| |
| − | rotkit.mutate("1HP1", chain="A", resi=308, target="CYS", mutframe=1)
| |
| − | cmd.refresh()
| |
| − | rotkit.mutate("1HP1", chain="A", resi=513, target="CYS", mutframe=1)
| |
| − | cmd.refresh()
| |
| − |
| |
| − | ##### Create simulated dye movement atoms
| |
| − | pseudoatom Donor, vdw=0.5
| |
| − | pseudoatom Acceptor, vdw=0.5
| |
| − | show spheres, Donor or Acceptor
| |
| − | rotkit.toline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name SG","Donor","Donor",15.0)
| |
| − | rotkit.toline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name SG","Acceptor","Acceptor",15.0)
| |
| − |
| |
| − | python
| |
| − | Dye_ang_incr = 6
| |
| − | Donor_angle_range = range(0,359,Dye_ang_incr)
| |
| − | Acceptor_angle_range = range(0,359,Dye_ang_incr)
| |
| − | nrstates = len(Donor_angle_range)+1
| |
| − | Donor_states = 1
| |
| − | Acceptor_states = 1
| |
| − | for Donor_angle in Donor_angle_range:
| |
| − | Donor_states += 1
| |
| − | Donor_angle_name="Donor_%s"%(Donor_angle)
| |
| − | cmd.create(Donor_angle_name,"Donor")
| |
| − | rotkit.rotateline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name CB",Donor_angle,Donor_angle_name)
| |
| − | # Save it as states in Donor
| |
| − | cmd.create("Donor",Donor_angle_name,1,Donor_states)
| |
| − | cmd.create("Donor",Donor_angle_name,1,2*nrstates-Donor_states)
| |
| − | cmd.group("All_Donors",Donor_angle_name)
| |
| − | for Acceptor_angle in Acceptor_angle_range:
| |
| − | Acceptor_states += 1
| |
| − | Acceptor_angle_name="Acceptor_%s"%(Acceptor_angle)
| |
| − | cmd.create(Acceptor_angle_name,"Acceptor")
| |
| − | rotkit.rotateline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name CB",Acceptor_angle,Acceptor_angle_name)
| |
| − | # Save it as states in Acceptor
| |
| − | cmd.create("Acceptor",Acceptor_angle_name,1,Acceptor_states)
| |
| − | cmd.create("Acceptor",Acceptor_angle_name,1,2*nrstates-Acceptor_states)
| |
| − | cmd.group("All_Acceptors",Acceptor_angle_name)
| |
| − | python end | |
| − | disable All_Donors
| |
| − | disable All_Acceptors
| |
| − | cmd.create("Donor","All_Donors",1,1)
| |
| − | cmd.create("Acceptor","All_Acceptors",1,1)
| |
| − | mplay
| |
| − | </syntaxhighlight>
| |
| | | | |
| | === Example 3 - Create distance distribution histogram === | | === Example 3 - Create distance distribution histogram === |
| − | <syntaxhighlight lang="python">
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_3.pml}} |
| − | reinitialize
| + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_3.pml" highlight="python" /> |
| − | #You need to make sure you are in the right dir, since we are going to make some datafiles
| |
| − | cd /home/tlinnet/test
| |
| − | import rotkit
| |
| − | | |
| − | fetch 1HP1, async=0
| |
| − | show_as cartoon, 1HP1
| |
| − | show_as sticks, 1HP1 and resn ATP
| |
| − | set auto_zoom, off
| |
| − |
| |
| − | ####################### Create rotate states of dye atoms ###################
| |
| − | ##### First mutate, the mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
| |
| − | rotkit.mutate("1HP1", chain="A", resi=308, target="CYS", mutframe=1)
| |
| − | cmd.refresh()
| |
| − | rotkit.mutate("1HP1", chain="A", resi=513, target="CYS", mutframe=1)
| |
| − | cmd.refresh()
| |
| − |
| |
| − | ##### Create simulated dye movement atoms
| |
| − | pseudoatom Donor, vdw=0.5
| |
| − | pseudoatom Acceptor, vdw=0.5
| |
| − | show spheres, Donor or Acceptor
| |
| − | rotkit.toline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name SG","Donor","Donor",15.0)
| |
| − | rotkit.toline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name SG","Acceptor","Acceptor",15.0)
| |
| − |
| |
| − | python
| |
| − | Dye_ang_incr = 6
| |
| − | Donor_angle_range = range(0,359,Dye_ang_incr)
| |
| − | Acceptor_angle_range = range(0,359,Dye_ang_incr)
| |
| − | Donor_names = []
| |
| − | Acceptor_names = []
| |
| − | for Donor_angle in Donor_angle_range:
| |
| − | Donor_angle_name="Donor_%s"%(Donor_angle)
| |
| − | Donor_names.append([Donor_angle,Donor_angle_name])
| |
| − | cmd.create(Donor_angle_name,"Donor")
| |
| − | rotkit.rotateline("1HP1 and resi 308 and name CA","1HP1 and resi 308 and name CB",Donor_angle,Donor_angle_name)
| |
| − | cmd.group("All_Donors",Donor_angle_name)
| |
| − | for Acceptor_angle in Acceptor_angle_range:
| |
| − | Acceptor_angle_name="Acceptor_%s"%(Acceptor_angle)
| |
| − | Acceptor_names.append([Acceptor_angle,Acceptor_angle_name])
| |
| − | cmd.create(Acceptor_angle_name,"Acceptor")
| |
| − | rotkit.rotateline("1HP1 and resi 513 and name CA","1HP1 and resi 513 and name CB",Acceptor_angle,Acceptor_angle_name)
| |
| − | cmd.group("All_Acceptors",Acceptor_angle_name)
| |
| − | python end
| |
| − | disable All_Donors
| |
| − | disable All_Acceptors
| |
| − | cmd.create("Donor","All_Donors")
| |
| − | cmd.create("Acceptor","All_Acceptors")
| |
| − | cmd.refresh()
| |
| − | | |
| − | # Make a distribution for the Open case
| |
| − | Don_Acc_distribution = []
| |
| − | python
| |
| − | for Don in Donor_names:
| |
| − | for Acc in Acceptor_names:
| |
| − | distname = "%s_%s"%(Don[1],Acc[1])
| |
| − | distance = cmd.dist(distname,Don[1],Acc[1])
| |
| − | Don_Acc_distribution.append([Don[0], Acc[1], distance])
| |
| − | cmd.delete(distname)
| |
| − | python end
| |
| − | Newdir=rotkit.createdirs("results_rotkit")
| |
| − | os.chdir(Newdir)
| |
| − | rotkit.makehistogram(Don_Acc_distribution,dataname="Don_Acc_Open",datalistindex=2,nrbins=100,binrange=[0,0])
| |
| − | | |
| − | # Make a distribution for angle range
| |
| − | cmd.create("Acceptor_rot","All_Acceptors")
| |
| − | python
| |
| − | ang_incr = 1
| |
| − | anglerange = range(2,98,ang_incr)
| |
| − | nrstates = len(anglerange)+1
| |
| − | states = 1
| |
| − | for angle in anglerange:
| |
| − | states += 1
| |
| − | rot_Acceptor = "Acceptor_rot_%s"%angle
| |
| − | cmd.create(rot_Acceptor,"Acceptor_rot")
| |
| − | rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",-(angle-1),rot_Acceptor)
| |
| − | cmd.create("Acceptor_rot",rot_Acceptor,1,states)
| |
| − | cmd.create("Acceptor_rot",rot_Acceptor,1,2*nrstates-states)
| |
| − | cmd.delete(rot_Acceptor)
| |
| − | for Acc in Acceptor_names:
| |
| − | rotkit.rotateline("/1HP1//A/423/CG1","/1HP1//A/477/C",(angle-1),Acc[1])
| |
| − | python end
| |
| − | </syntaxhighlight>
| |
| | | | |
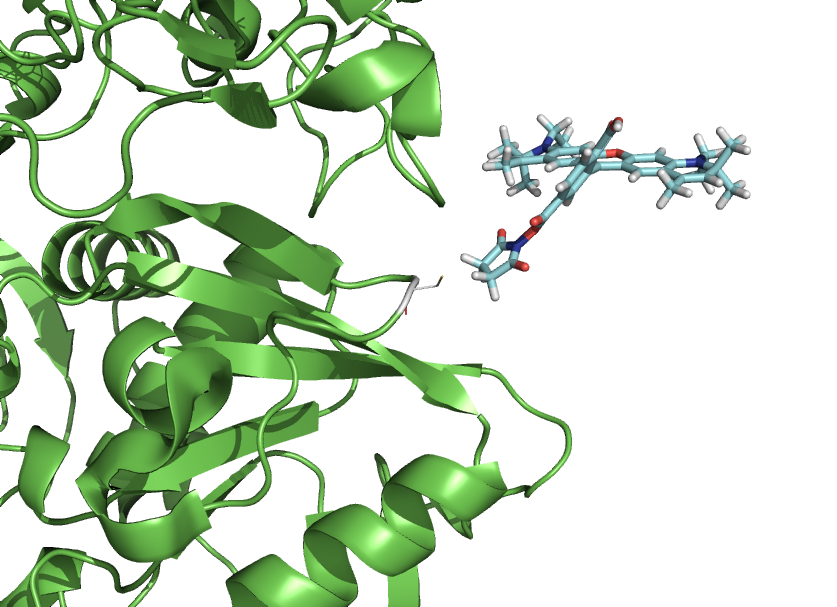
| | === Example 4 - A tutorial file === | | === Example 4 - A tutorial file === |
| − |
| |
| | To understand how the functions works, read through the tutorial. Hash/Unhash "##" each step at the time to see the effect. | | To understand how the functions works, read through the tutorial. Hash/Unhash "##" each step at the time to see the effect. |
| | + | To be able to follow the tutorial, you need the dye molecule, which is loaded from the Pymol-script-repository. |
| | | | |
| − | Make a test dir: '''/home/tlinnet/test''' and make a '''tutorial.pml''' with the commands.
| + | {{Template:PymolScriptRepoDownload|examples/rotkit_4.pml}} |
| − | | + | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/rotkit_4.pml" highlight="python" /> |
| − | To be able to follow the tutorial, you need the dye molecule.
| |
| − | Right clicking the following link here -> Save as: Atto590.pdb in the directory with the tutorial.pml file
| |
| − | https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/files_for_examples/Atto590.pdb
| |
| − | | |
| − | <syntaxhighlight lang="python">
| |
| − | reinitialize
| |
| − | cd /home/tlinnet/test
| |
| − | import rotkit
| |
| − |
| |
| − | fetch 1HP1, async=0
| |
| − | load Atto590.pdb
| |
| − | # Make sure everything is loaded before we continue
| |
| − | cmd.refresh()
| |
| − | | |
| − | ### Get the names of the loaded objects
| |
| − | protname = cmd.get_names()[0]
| |
| − | molname = cmd.get_names()[1]
| |
| − | | |
| − | ### Make the names we are going to use
| |
| − | protselectCB="%s and resi 308 and name CB"%protname
| |
| − | protnameselectCB="K308CB"
| |
| − | protselectCA="%s and resi 308 and name CA"%protname
| |
| − | protnameselectCA="K308CA"
| |
| − | molselect13="%s and id 13"%molname
| |
| − | molnameselect13="dyeatom13"
| |
| − | molselect12="%s and id 12"%molname
| |
| − | molnameselect12="dyeatom12"
| |
| − |
| |
| − | ### Make some selections
| |
| − | cmd.select("%s"%protnameselectCB,"%s"%protselectCB)
| |
| − | cmd.select("%s"%protnameselectCA,"%s"%protselectCA)
| |
| − | cmd.select("%s"%molnameselect13,"%s"%molselect13)
| |
| − | cmd.label("%s"%molnameselect13,"13")
| |
| − | cmd.select("%s"%molnameselect12,"%s"%molselect12)
| |
| − | cmd.label("%s"%molnameselect12,"12")
| |
| − | | |
| − | ### Make nice representations
| |
| − | cmd.show_as("cartoon","%s"%protname)
| |
| − | cmd.show("sticks","byres %s"%protnameselectCB)
| |
| − |
| |
| − | ##### PART I: Use of functions #####
| |
| − | ### This view will take you to the first part
| |
| − | set_view (\
| |
| − | 0.377224118, 0.880101919, -0.288305759,\
| |
| − | 0.661396861, -0.473919988, -0.581338286,\
| |
| − | -0.648268998, 0.028612033, -0.760871351,\
| |
| − | 0.000000000, 0.000000000, -56.408561707,\
| |
| − | 19.480533600, 34.572898865, 6.978204727,\
| |
| − | 46.615653992, 66.201446533, -20.000001907 )
| |
| − |
| |
| − | #### Just unhash each part for itself, as you continue through
| |
| − | ### To print a objects TTT matrix in a readable format
| |
| − | rotkit.printMat(cmd.get_object_matrix(molname))
| |
| − |
| |
| − | ##### We want to move the dye to a desired location, and rotate it to a view we desire
| |
| − | ##### First get the vector bewteen the dyeatom and the protein atom
| |
| − | diffvector = rotkit.vector("%s"%molselect13,"%s"%protnameselectCB)
| |
| − | ##### Then move the dye
| |
| − | move = rotkit.transmat(diffvector)
| |
| − | ##### print the matrix for fun
| |
| − | rotkit.printMat(move)
| |
| − | ##### Move the dye
| |
| − | cmd.transform_selection("%s"%molname,move)
| |
| − | | |
| − | ##### Now we want to displace the dye in the CA-CB bond direction
| |
| − | ##### First find the vector/direction to displace it in. From A -> B
| |
| − | diffvector = rotkit.vector("%s"%protnameselectCA,"%s"%protnameselectCB)
| |
| − | ##### Make the vector so its lenth is equal 1
| |
| − | uvector = rotkit.unitvector(diffvector)[0]
| |
| − | ##### Make the move translation matrix, and we multiply the matrix with 3, so it moves 3 Angstrom
| |
| − | move = rotkit.transmat(uvector,3)
| |
| − | ##### Print the matrix
| |
| − | rotkit.printMat(move)
| |
| − | ##### Displace it in the CA-CB direction
| |
| − | cmd.transform_selection("%s"%molname,move)
| |
| − | | |
| − | ##### Now we want to rotate it a single time. We convert 40 degress to radians
| |
| − | ##### The input is the angle, the line to rotate around, and a point where the line goes through
| |
| − | CBxyz = rotkit.getxyz("%s"%protnameselectCB)[0]
| |
| − | rmat = rotkit.rotmat(rotkit.radangle(40),uvector,CBxyz)
| |
| − | rotkit.printMat(rmat)
| |
| − | ##### Copy paste this line into pymol to see it manually
| |
| − | cmd.transform_selection("%s"%molname,rmat)
| |
| − | | |
| − | ##### We are not quite satisfied, we want to rotate it around its own bond
| |
| − | ##### So we rotate in around its own 13 -> 12 bonds
| |
| − | diffvector = rotkit.vector("%s"%molnameselect13,"%s"%molnameselect12)
| |
| − | uvector = rotkit.unitvector(diffvector)[0]
| |
| − | xyz12 = rotkit.getxyz("%s"%molnameselect12)[0]
| |
| − | rmat = rotkit.rotmat(rotkit.radangle(10),uvector,xyz12)
| |
| − | ##### Copy paste this line into pymol to see it manually
| |
| − | cmd.transform_selection("%s"%molname,rmat)
| |
| − | | |
| − | ##### Now, lets make a function that collects all these call in one function
| |
| − | ##### We only want to define two positions that defines the line, the angle and the object to rotate
| |
| − | rotkit.rotateline("%s"%molnameselect13,"%s"%molnameselect12,180,"%s"%molname)
| |
| − | ##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
| |
| − | print("rotateline Pos1=%s, Pos2=%s, degangle=15, molecule=%s"%(molnameselect13, molnameselect12, molname))
| |
| − | | |
| − | ##### To illustate best, we create som copies of the dye
| |
| − | python
| |
| − | anglerange = range(90,360,90)
| |
| − | for angle in anglerange:
| |
| − | ### Make a suitable name for the new molecule
| |
| − | molanglename="%s%s"%(molname,angle)
| |
| − | ### Now make a copy
| |
| − | cmd.create(molanglename,molname)
| |
| − | cmd.label("%s and id 12"%molanglename,"12")
| |
| − | cmd.label("%s and id 13"%molanglename,"13")
| |
| − | ### Rotate the copy
| |
| − | rotkit.rotateline("%s"%protnameselectCB,"%s"%molnameselect13,angle,"%s"%molanglename)
| |
| − | python end | |
| − | | |
| − | | |
| − | ####### End of PART I ####
| |
| − | ####### PART II: More advanced functions #####
| |
| − | ##### This view will take you to the second part
| |
| − | set_view (\
| |
| − | 0.723298192, 0.467510879, 0.508201897,\
| |
| − | 0.371686131, -0.883831143, 0.284063697,\
| |
| − | 0.581970334, -0.016570913, -0.813038886,\
| |
| − | 0.000000000, 0.000000000, -76.609786987,\
| |
| − | 11.790571213, 64.992294312, 20.803859711,\
| |
| − | -31.181428909, 184.401092529, -20.000001907 )
| |
| − | | |
| − | ##### We can fast mutate a protein. frame 1 is the most probable mutation
| |
| − | rotkit.mutate(protname, chain="A", resi=513, target="CYS", mutframe=1)
| |
| − | ##### The mutate functions take 0.2 seconds, so we put in a refesh command to wait for everything is done
| |
| − | cmd.refresh()
| |
| − | ##### This is made as a pymol command as well. I first print the names that we should write manually in the consol
| |
| − | print("mutate %s, chain=%s, resi=%s, target=CYS, mutframe=1"%(protname, "A", 515))
| |
| − | | |
| − | ##### We now make some selections for this mutation
| |
| − | protselectCBcys="%s and resi 513 and name CB"%protname
| |
| − | protnameselectCBcys="P513C_CB"
| |
| − | protselectCAcys="%s and resi 513 and name CA"%protname
| |
| − | protnameselectCAcys="P513C_CA"
| |
| − | cmd.select("%s"%protnameselectCBcys,"%s"%protselectCBcys)
| |
| − | cmd.select("%s"%protnameselectCAcys,"%s"%protselectCAcys)
| |
| − | | |
| − | ##### Now, lets make a function that collects all the commands to put on an atom on the same line defined by two points
| |
| − | ##### The input is the two points that define the line, the atom of a molecule to be put on the line, and the distance to move
| |
| − | rotkit.toline(protnameselectCAcys,protnameselectCBcys,molnameselect13,molname,3)
| |
| − | rotkit.rotateline(protnameselectCAcys,protnameselectCBcys,180,molname)
| |
| − | rotkit.rotateline(molnameselect13,molnameselect12,10,molname)
| |
| − | print("toline Pos1=%s, Pos2=%s, atom=%s, molecule=%s, dist=%s"%(protnameselectCAcys,protnameselectCBcys,molnameselect13,molname,3))
| |
| − | print("rotateline Pos1=%s, Pos2=%s, degangle=180, molecule=%s"%(protnameselectCAcys, protnameselectCBcys, molname))
| |
| − | print("rotateline Pos1=%s, Pos2=%s, degangle=10, molecule=%s"%(molnameselect13, molnameselect12, molname))
| |
| − | cmd.refresh()
| |
| − | ####### End of PART II ####
| |
| − | | |
| − | ####### Now we make a cross product ####
| |
| − | molselect14="%s and id 14"%molname
| |
| − | molnameselect14="dyeatom14"
| |
| − | cmd.select("%s"%molnameselect14,"%s"%molselect14)
| |
| − | cmd.label("%s"%molnameselect14,"14")
| |
| − | | |
| − | cross = rotkit.crossprod(rotkit.vector(molselect13,molselect12),rotkit.vector(molselect13,molselect14))
| |
| − | unity_cross = rotkit.unitvector(cross)[0]
| |
| − | point_cross = rotkit.crosspoint(molselect13,cross)
| |
| − | rotkit.rotateline(molnameselect13,point_cross,180,molname)
| |
| − | print("rotateline Pos1=%s, Pos2=%s, degangle=10, molecule=%s"%(molnameselect13, pcross, molname))
| |
| − | </syntaxhighlight>
| |
| | | | |
| | [[Category:Script_Library]] | | [[Category:Script_Library]] |
| | [[Category:ObjSel_Scripts]] | | [[Category:ObjSel_Scripts]] |
| | + | [[Category:Pymol-script-repo]] |