This is a read-only mirror of pymolwiki.org
Difference between revisions of "Mole"
(Mole software) |
m (3 revisions) |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ====There is a new version of MOLE [[MOLE 2.0: advanced approach for analysis of biomacromolecular channels|check it out]]!==== | ||
| + | |||
| + | |||
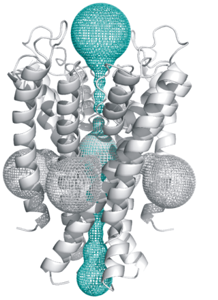
[http://mole.chemi.muni.cz MOLE] is an universal toolkit for rapid and fully automated location and characterization of channels, tunnels and pores in molecular structures. The core of MOLE algorithm is a Dijsktra path search algorithm, which is applied to a Voronoi mesh. MOLE is a powerful software (overcomming some limitations of [[Caver]] tool) for exploring large molecular channels, complex networks of channels and molecular dynamics trajectories (AMBER ascii traj and parm7 are supported) in which analysis of a large number of snapshots is required. | [http://mole.chemi.muni.cz MOLE] is an universal toolkit for rapid and fully automated location and characterization of channels, tunnels and pores in molecular structures. The core of MOLE algorithm is a Dijsktra path search algorithm, which is applied to a Voronoi mesh. MOLE is a powerful software (overcomming some limitations of [[Caver]] tool) for exploring large molecular channels, complex networks of channels and molecular dynamics trajectories (AMBER ascii traj and parm7 are supported) in which analysis of a large number of snapshots is required. | ||
| Line 14: | Line 17: | ||
[[Category:Biochemical_Properties]] | [[Category:Biochemical_Properties]] | ||
[[Category:Plugins]] | [[Category:Plugins]] | ||
| + | [[Category:Pymol-script-repo]] | ||
Latest revision as of 01:53, 28 March 2014
There is a new version of MOLE check it out!
MOLE is an universal toolkit for rapid and fully automated location and characterization of channels, tunnels and pores in molecular structures. The core of MOLE algorithm is a Dijsktra path search algorithm, which is applied to a Voronoi mesh. MOLE is a powerful software (overcomming some limitations of Caver tool) for exploring large molecular channels, complex networks of channels and molecular dynamics trajectories (AMBER ascii traj and parm7 are supported) in which analysis of a large number of snapshots is required.
Program is available as standalone application for Linux(Unix), MacOS and Windows. Plugin for pyMol enables calling the MOLE program via an user friendly interface and allows results visualization. The on-line interface provides another interactive and easy-to-use interface to analyze molecular channels.
Tutorial is also available.
References:
Petrek M., Kosinova P., Koca J., Otyepka M.: MOLE: A Voronoi Diagram-Based Explorer of Molecular Channels, Pores, and Tunnels. Structure 2007, 15, 1357-1363