This is a read-only mirror of pymolwiki.org
Difference between revisions of "3DNA"
(→3DNA) |
m (1 revision) |
||
| (38 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | '' '''Update:''' See also the [[dssr_block]] wrapper script for '''x3dna-dssr'''.'' | |
| + | 3DNA[http://x3dna.org/] provides a ruby script (<b>blocview</b>) which produces Calladine-Drew style block representations based on the standard reference frame for nucleic acids. The produced files are in raster3D format, and are also processed by molscript. Therefore to produce the following representations you will need to install: | ||
| + | - 3DNA [http://x3dna.org/] | ||
| + | - Raster 3D [http://skuld.bmsc.washington.edu/raster3d/raster3d.html] | ||
| − | + | - Molscript [http://www.avatar.se/molscript/download.html] | |
| − | - | + | Once you have the previous software installed, and the path to their binaries correctly configured, you can get the block representation for any nucleic acid. The next example shows how to do it for tRNA (PDB:ID 1ehz) |
| − | - | + | |
| − | - | + | == Invoking == |
| + | <source lang="bash"> | ||
| + | pdb_get.py 1ehz | ||
| + | blocview -o 1ehz.pdb | ||
| + | </source> | ||
| + | |||
| + | The pdb_get python script comes from [http://pldserver1.biochem.queensu.ca/~rlc/work/scripts/pdb_get.py]Dr. Robert Campbell's website. | ||
| + | |||
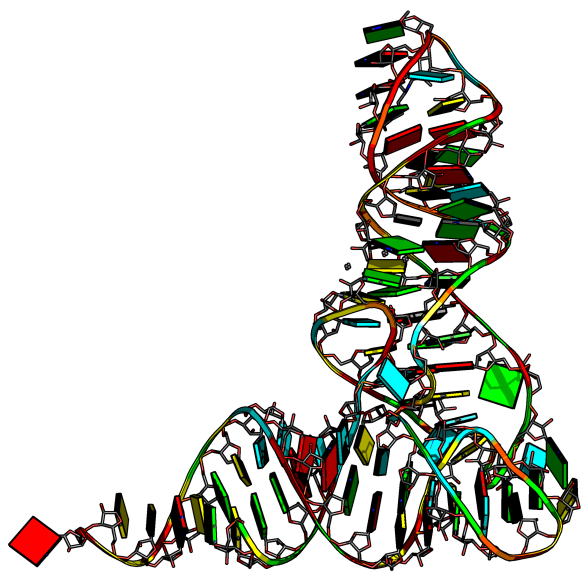
| + | Once blocview is run it will generate various r3d files which can be combined with the original pdb files to produce the following image: | ||
| + | |||
| + | == Example 1 == | ||
| + | <gallery> | ||
| + | Image:trna.png|Using tb.pdb and t2.r3d | ||
| + | </gallery> | ||
| + | |||
| + | To generate the previous image the following script was used: | ||
| + | trna.pml | ||
| + | == Syntax == | ||
| + | <source lang="python"> | ||
| + | load tb.pdb | ||
| + | load t2.r3d | ||
| + | bg_color white | ||
| + | hide lines | ||
| + | zoom *,-5 | ||
| + | set ray_trace_mode, 3 | ||
| + | set ray_trace_fog, 0 | ||
| + | set ray_shadows, 0 | ||
| + | set orthoscopic, 1 | ||
| + | set antialias, 5 | ||
| + | set valence, 1 | ||
| + | util.cba(29) | ||
| + | color grey, (elem C) | ||
| + | cartoon arrow | ||
| + | set cartoon_ladder_mode, 0 | ||
| + | set cartoon_rect_width, 0.2 | ||
| + | set cartoon_rect_length, 0.5 | ||
| + | show cartoon | ||
| + | set stick_radius, 0.14 | ||
| + | show sticks | ||
| + | set_view (\ | ||
| + | 0.680474579, -0.153203458, -0.716576934,\ | ||
| + | 0.658882320, -0.300013036, 0.689829707,\ | ||
| + | -0.320666909, -0.941552401, -0.103208199,\ | ||
| + | -0.000084338, 0.000022471, -263.030426025,\ | ||
| + | 57.723434448, 45.338260651, 20.895099640,\ | ||
| + | 218.710235596, 307.348541260, 1.000000000 ) | ||
| + | set cartoon_color, green, resn G | ||
| + | set cartoon_color, yellow, resn C | ||
| + | set cartoon_color, red, resn A | ||
| + | set cartoon_color, cyan, resn U | ||
| + | ray 1024,768 | ||
| + | png trna.png | ||
| + | quit | ||
| + | </source> | ||
| + | |||
| + | == Example 2 == | ||
| + | |||
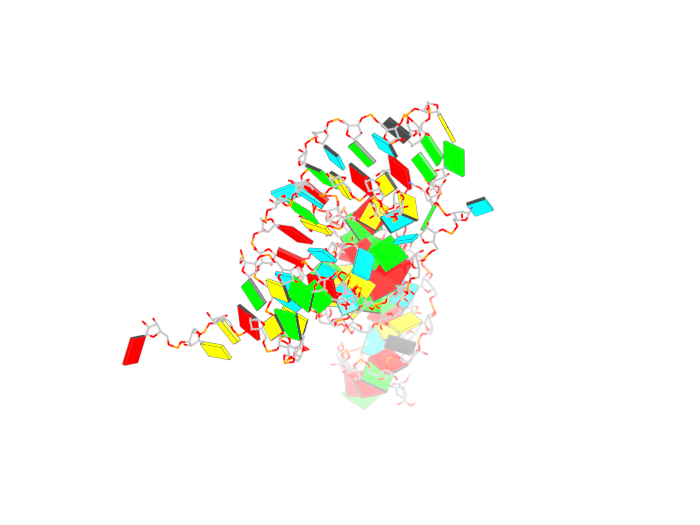
| + | An automatically generated pymol ray traced image can also be obtained by running <b>blocview</b> from 3DNA v. 2.0 directly. Using the following command: | ||
| + | |||
| + | <pre> | ||
| + | blocview -o -t=100 1ehz.pdb | ||
| + | </pre> | ||
| + | |||
| + | And the result is: | ||
| + | |||
| + | <gallery> | ||
| + | Image:X3dna_r3d_pymol.png | ||
| + | </gallery> | ||
| + | |||
| + | |||
| + | == More == | ||
| + | |||
| + | For more examples of figures obtained using 3DNA and pymol follow the next link: | ||
| + | [[http://mesguerra.org/render/render.htm]] | ||
| + | |||
| + | [[Category:Nucleic_Acids]] | ||
Latest revision as of 05:26, 8 February 2016
Update: See also the dssr_block wrapper script for x3dna-dssr.
3DNA[1] provides a ruby script (blocview) which produces Calladine-Drew style block representations based on the standard reference frame for nucleic acids. The produced files are in raster3D format, and are also processed by molscript. Therefore to produce the following representations you will need to install:
- 3DNA [2]
- Raster 3D [3]
- Molscript [4]
Once you have the previous software installed, and the path to their binaries correctly configured, you can get the block representation for any nucleic acid. The next example shows how to do it for tRNA (PDB:ID 1ehz)
Invoking
pdb_get.py 1ehz
blocview -o 1ehz.pdb
The pdb_get python script comes from [5]Dr. Robert Campbell's website.
Once blocview is run it will generate various r3d files which can be combined with the original pdb files to produce the following image:
Example 1
To generate the previous image the following script was used: trna.pml
Syntax
load tb.pdb
load t2.r3d
bg_color white
hide lines
zoom *,-5
set ray_trace_mode, 3
set ray_trace_fog, 0
set ray_shadows, 0
set orthoscopic, 1
set antialias, 5
set valence, 1
util.cba(29)
color grey, (elem C)
cartoon arrow
set cartoon_ladder_mode, 0
set cartoon_rect_width, 0.2
set cartoon_rect_length, 0.5
show cartoon
set stick_radius, 0.14
show sticks
set_view (\
0.680474579, -0.153203458, -0.716576934,\
0.658882320, -0.300013036, 0.689829707,\
-0.320666909, -0.941552401, -0.103208199,\
-0.000084338, 0.000022471, -263.030426025,\
57.723434448, 45.338260651, 20.895099640,\
218.710235596, 307.348541260, 1.000000000 )
set cartoon_color, green, resn G
set cartoon_color, yellow, resn C
set cartoon_color, red, resn A
set cartoon_color, cyan, resn U
ray 1024,768
png trna.png
quit
Example 2
An automatically generated pymol ray traced image can also be obtained by running blocview from 3DNA v. 2.0 directly. Using the following command:
blocview -o -t=100 1ehz.pdb
And the result is:
More
For more examples of figures obtained using 3DNA and pymol follow the next link: [[6]]