This is a read-only mirror of pymolwiki.org
Difference between revisions of "Builder"
m (Incentive PyMOL 2.3) |
m (2 revisions) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 36: | Line 36: | ||
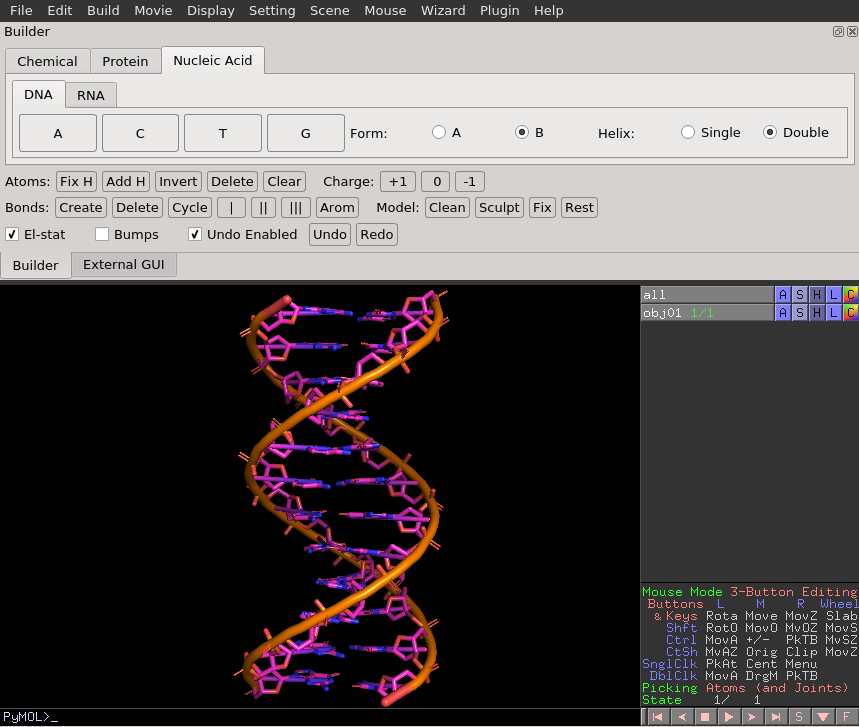
Here is an example finished product from the DNA builder. | Here is an example finished product from the DNA builder. | ||
| + | |||
| + | ''New in Incentive PyMOL 2.5'' | ||
| + | |||
| + | As of Incentive PyMOL 2.5, builder can attach nucleic acids to existing structures. Double stranded structures are automatically detected based on distance cutoff and base pairing. Additionally, a phosphate group will be automatically added to the 5' end if missing from the structure. | ||
| + | |||
| + | The "Residue: Remove" button will remove the residue currently selected based on PyMOL's Selection Algebra. This means the entire residue of the atom selected will be removed. This works for both proteins and nucleic acids. | ||
| + | [[File:Remove-Residue-Button.png]] | ||
[[Category:Builder]] | [[Category:Builder]] | ||
Latest revision as of 03:21, 22 June 2021
Overview
The Builder is a PyMOL GUI menu that allows you to easily build up structures by hand from various elements like atoms, fragments, rings, amino acids, etc. You can also assign charge, secondary structure, etc.
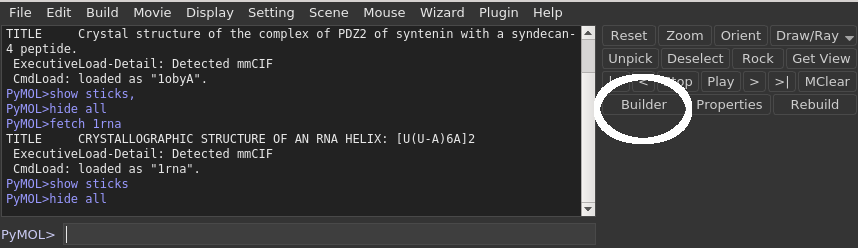
To access the Builder simply select the "Builder" option from the PyMOL GUI (see images).
Nucleic Acid Builder
As of Incentive PyMOL 2.3, building can also be done with nucleic acids.
Click the 'Builder' button in the top-right panel.
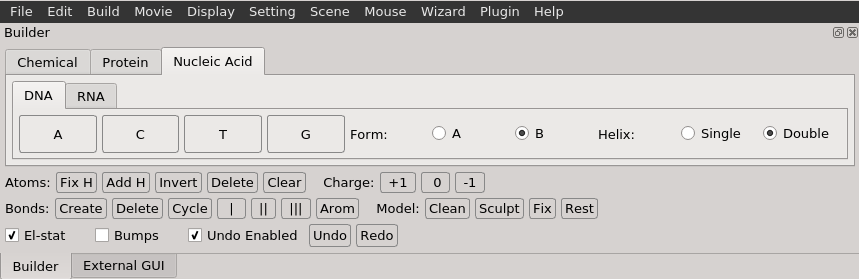
Once selecting "Nucleic Acids", you will presented options for building either DNA or RNA. For DNA, you can specify the custom DNA's form (A-DNA or B-DNA) and whether the DNA should be single- or double-stranded.
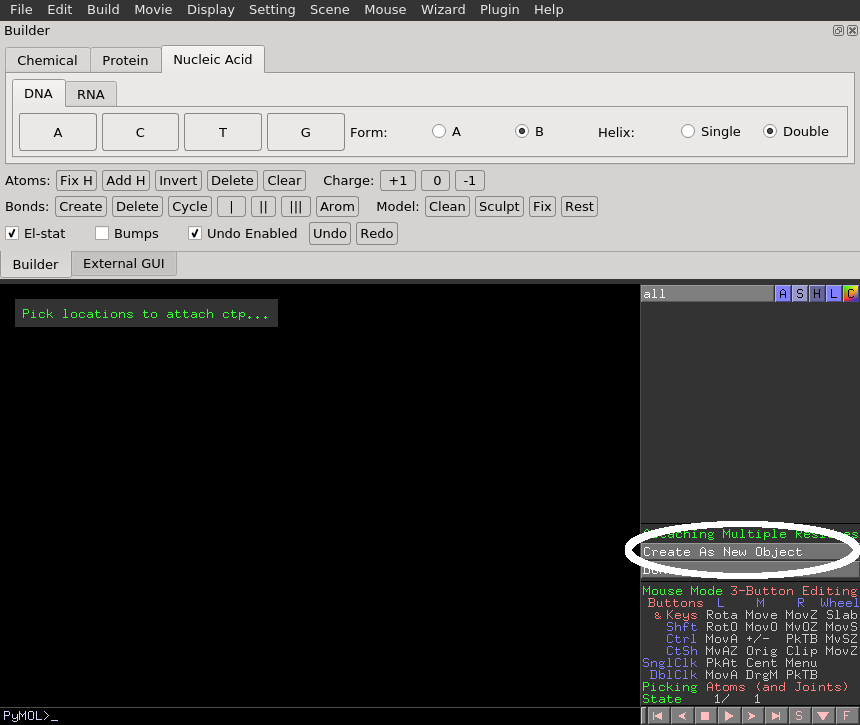
When ready, select the base type for the nucleic acid by clicking on the button with its specified one-letter code (e.g. A, T, U, C, or G) and select "Create As New Object".
To extend the DNA, select a terminal 5' phosphate or 3' oxygen.
You can see the change in the nucleic acid's sequence in real-time (Display-> Sequence). For double-stranded DNA, positive residue indices are base-paired to its negative counterpart (5 base-paired with -5, 12 base-paired with -12, etc...).
Once finished, click the "Done" button.
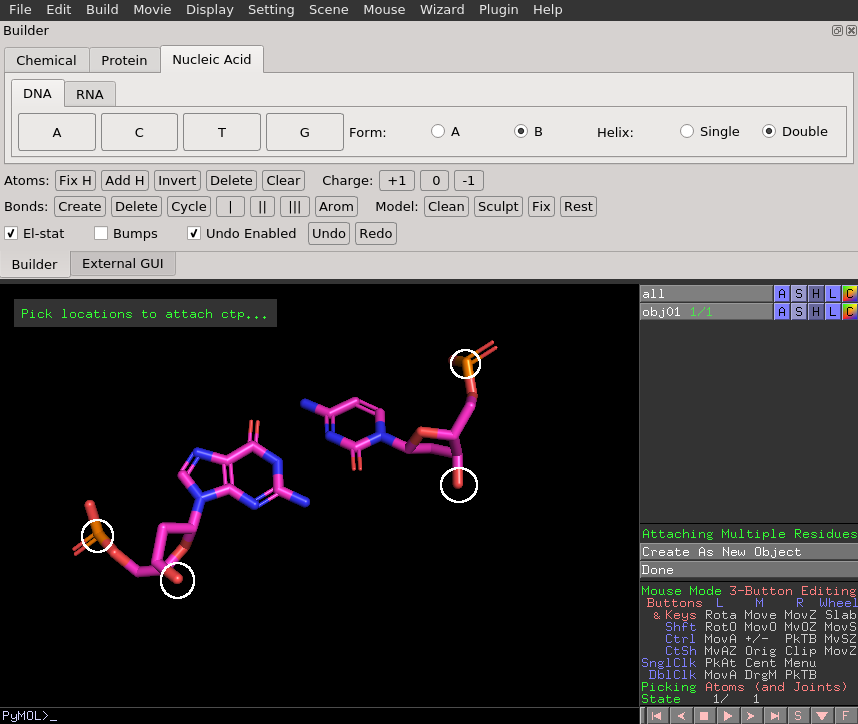
Here is an example finished product from the DNA builder.
New in Incentive PyMOL 2.5
As of Incentive PyMOL 2.5, builder can attach nucleic acids to existing structures. Double stranded structures are automatically detected based on distance cutoff and base pairing. Additionally, a phosphate group will be automatically added to the 5' end if missing from the structure.
The "Residue: Remove" button will remove the residue currently selected based on PyMOL's Selection Algebra. This means the entire residue of the atom selected will be removed. This works for both proteins and nucleic acids. File:Remove-Residue-Button.png