This is a read-only mirror of pymolwiki.org
Difference between revisions of "Selection Algebra"
m (1 revision) |
m (4 revisions) |
||
| (21 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
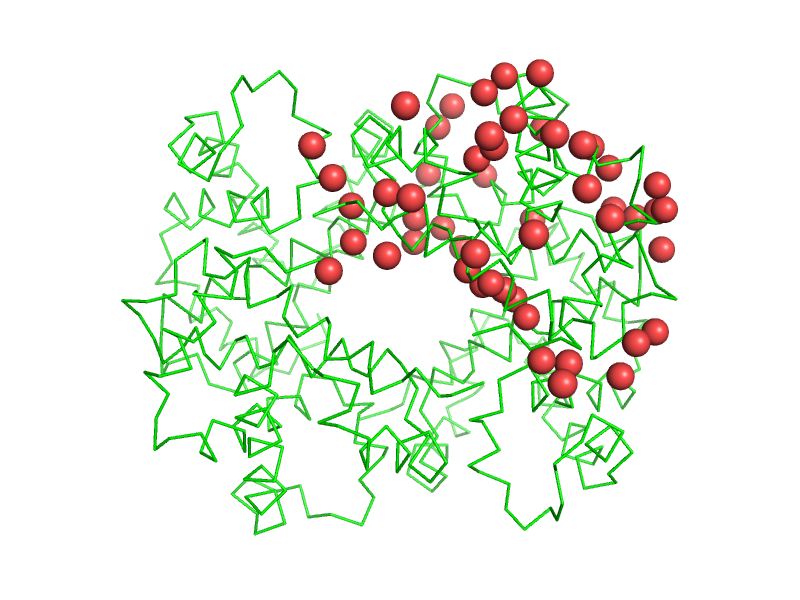
| − | + | [[Image:solvent-and-chain-A.png|thumb|240px|show spheres, solvent and chain A]] | |
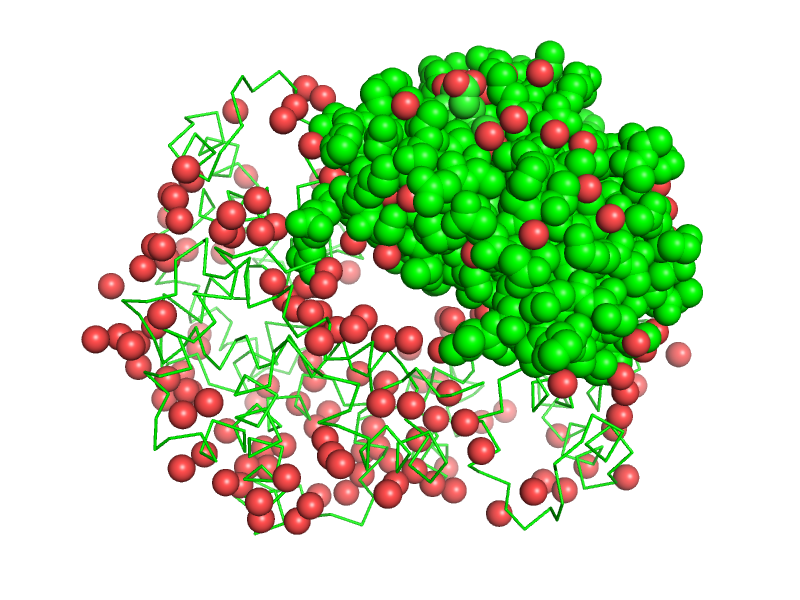
| + | [[Image:solvent-or-chain-A.png|thumb|240px|show spheres, solvent or chain A]] | ||
| − | + | PyMOL's selection language allows to select atoms based on identifiers and properties. | |
| + | Many commands (like [[color]], [[show]], etc.) take an atom selection argument to only operate on a subset of all atoms in the scene. | ||
| + | Example: | ||
| − | + | PyMOL>show spheres, solvent and chain A | |
| + | |||
| + | Selections can be made more precise or inclusive by combining them with logical operators, including the boolean '''and''', '''or''', and '''not'''. The boolean '''and''' selects only those items that have both (or all) of the named properties, and the boolean '''or''' selects items that have either (or any) of them. | ||
== Selection Operator/Modifier Table == | == Selection Operator/Modifier Table == | ||
Selection operators and modifiers are listed below. The dummy variables ''s1'' and ''s2'' stand for selection-expressions such as "chain a" or "hydro." | Selection operators and modifiers are listed below. The dummy variables ''s1'' and ''s2'' stand for selection-expressions such as "chain a" or "hydro." | ||
| − | + | {| class="wikitable" | |
| − | + | |- | |
| − | + | ! Operator | |
| − | + | ! Aliases | |
| − | + | ! Description | |
| − | < | + | |- |
| − | < | + | !colspan="3" style="text-align:left"| Generic |
| − | + | |- | |
| − | < | + | | all || * |
| − | + | | All atoms currently loaded into PyMOL | |
| − | < | + | |- |
| − | + | | none || | |
| − | < | + | | Empty selection |
| − | < | + | |- |
| − | < | + | | enabled || |
| − | < | + | | Atoms from enabled objects |
| − | + | |- | |
| − | + | !colspan="3" style="text-align:left"| Named selections | |
| − | + | |- | |
| − | < | + | | <span style="color: #999">sele</span> || |
| − | + | | Named selection or object "sele", but only if it doesn't conflict with the name of another operator | |
| − | < | + | |- |
| − | < | + | | %<span style="color: #999">sele</span> || |
| − | < | + | | Named selection or object "sele" <span style="padding: 1px 4px; background-color: #fc3; border: 1px solid #ccc">Recommended, avoids ambiguity</span> |
| − | + | |- | |
| − | + | | ?<span style="color: #999">sele</span> || | |
| − | < | + | | Named selection or object "sele", or empty selection if "sele" doesn't exist |
| − | + | |- | |
| − | + | !colspan="3" style="text-align:left"| Logical | |
| − | + | |- | |
| − | < | + | | not <span style="color: #999">S1</span> || ! |
| − | + | | Inverts selection | |
| − | + | |- | |
| − | + | | <span style="color: #999">S1</span> and <span style="color: #999">S2</span> || & | |
| − | < | + | | Atoms included in both S1 and S2 |
| − | < | + | |- |
| − | < | + | | <span style="color: #999">S1</span> or <span style="color: #999">S2</span> || <nowiki>|</nowiki> |
| − | + | | Atoms included in either S1 or S2 | |
| − | < | + | |- |
| − | + | | <span style="color: #999">S1 S2</span> || | |
| − | + | | implicit '''or''' | |
| − | + | |- | |
| − | + | | <span style="color: #999">S1</span> and (<span style="color: #999">S2</span> or <span style="color: #999">S3</span>) || | |
| − | + | | Parentheses for evaluation order control | |
| − | + | |- | |
| − | + | | first <span style="color: #999">S1</span> || | |
| − | + | | First atom in S1 (single atom selection) | |
| − | < | + | |- |
| − | + | | last <span style="color: #999">S1</span> || | |
| − | < | + | | Last atom in S1 (single atom selection) |
| − | + | |- | |
| − | < | + | !colspan="3" style="text-align:left"| Identifiers <small style="font-weight: normal">(see also [[Selection Macros]])</small> |
| − | + | |- | |
| − | < | + | | model <span style="color: #999">1ubq</span> || m. |
| − | + | | Atoms from object "1ubq" | |
| − | + | |- | |
| − | < | + | | chain <span style="color: #999">C</span> || c. |
| − | + | | Chain identifier "C" | |
| − | + | |- | |
| − | + | | segi <span style="color: #999">S</span> || s. | |
| − | < | + | | Segment identifier "S" ('''label_asym_id''' from mmCIF) |
| − | < | + | |- |
| − | + | | resn <span style="color: #999">ALA</span> || r. | |
| − | < | + | | Residue name "ALA" |
| − | < | + | |- |
| − | < | + | | resi <span style="color: #999">100-200</span> || i. |
| − | + | | Residue identifier between 100 and 200 | |
| − | + | |- | |
| − | + | | name <span style="color: #999">CA</span> || n. | |
| − | + | | Atom name "CA" | |
| − | + | |- | |
| − | + | | alt <span style="color: #999">A</span> || | |
| − | + | | Alternate location "A" | |
| − | + | |- | |
| − | + | | index <span style="color: #999">123</span> || idx. | |
| − | + | | Internal per-object atom index (changes with [[sort|sorting]]) | |
| − | + | |- | |
| − | < | + | | id <span style="color: #999">123</span> || |
| − | + | | ID column from PDB file | |
| − | + | |- | |
| − | + | | rank <span style="color: #999">123</span> || | |
| − | + | | Per-object atom index at load time (see also [[retain_order]]) | |
| − | + | |- | |
| − | + | | pepseq <span style="color: #999">ACDEF</span> || ps. | |
| − | + | | Protein residue sequence with one-letter code "ACDEF" (see also [[FindSeq]]) | |
| − | + | |- | |
| − | + | | label <span style="color: #999">"Hello World"</span> || | |
| − | + | | Atoms with label "Hello World" ''(new in PyMOL 1.9)'' | |
| − | + | |- | |
| − | + | !colspan="3" style="text-align:left"| Identifier matching | |
| − | + | |- | |
| − | + | | <span style="color: #999">S1</span> in <span style="color: #999">S2</span> || | |
| − | + | | Atoms in S1 whose identifiers ''name, resi, resn, chain'' and ''segi'' '''all''' match atoms in S2 | |
| + | |- | ||
| + | | <span style="color: #999">S1</span> like <span style="color: #999">S2</span> || | ||
| + | | Atoms in S1 whose identifiers ''name'' and ''resi'' match atoms in S2 | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Entity expansion | ||
| + | |- | ||
| + | |colspan="3" style="background-color: #fc3"| <span id="weak-by">Important:</span> All "by"-operators have a '''weak priority''', so (byres S1 or S2) is actually identical to (byres (S1 or S2)) and '''not''' to ((byres S1) or S2) | ||
| + | |- | ||
| + | | byobject <span style="color: #999">S1</span> || | ||
| + | | Expands S1 to complete objects | ||
| + | |- | ||
| + | | bysegi <span style="color: #999">S1</span> || bs. | ||
| + | | Expands S1 to complete segments | ||
| + | |- | ||
| + | | bychain <span style="color: #999">S1</span> || bc. | ||
| + | | Expands S1 to complete chains | ||
| + | |- | ||
| + | | byres <span style="color: #999">S1</span> || br. | ||
| + | | Expands S1 to complete residues | ||
| + | |- | ||
| + | | bycalpha <span style="color: #999">S1</span> || bca. | ||
| + | | CA atoms of residues with at least one atom in S1 | ||
| + | |- | ||
| + | | bymolecule <span style="color: #999">S1</span> || bm. | ||
| + | | Expands S1 to complete molecules (connected with bonds) | ||
| + | |- | ||
| + | | byfragment <span style="color: #999">S1</span> || bf. | ||
| + | | | ||
| + | |- | ||
| + | | byring <span style="color: #999">S1</span> || | ||
| + | | All rings of size ≤ 7 which have at least one atom in S1 ''(new in PyMOL 1.8.2)'' | ||
| + | |- | ||
| + | | bycell <span style="color: #999">S1</span> || | ||
| + | | Expands selection to unit cell | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Bond expansion | ||
| + | |- | ||
| + | | bound_to <span style="color: #999">S1</span> || bto. | ||
| + | | Atoms directly bonded to S1, may include S1 | ||
| + | |- | ||
| + | | neighbor <span style="color: #999">S1</span> || nbr. | ||
| + | | Atoms directly bonded to S1, excludes S1 | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> extend <span style="color: #999">3</span> || xt. | ||
| + | | Expands S1 by 3 bonds connected to atoms in S1 | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Proximity <small style="font-weight: normal">(see also [[#Comparison of distance operators|comparison of distance operators]])</small> | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> within <span style="color: #999">12.3</span> of <span style="color: #999">S2</span> || w. | ||
| + | | Atoms in S1 that are within 12.3 Angstroms of any atom in S2 | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> around <span style="color: #999">12.3</span> || a. | ||
| + | | Atoms with centers within 12.3 Angstroms of the center of any atom in S1 | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> expand <span style="color: #999">12.3</span> || x. | ||
| + | | Expands S1 by atoms within 12.3 Angstroms of the center of any atom in S1 | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> gap <span style="color: #999">1.2</span> || | ||
| + | | Atoms whose VDW radii are separated from the VDW radii of S1 by a minimum of 1.2 Angstroms. | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> near_to <span style="color: #999">12.3</span> of <span style="color: #999">S2</span> || nto. | ||
| + | | Same as ''within'', but excludes S2 from the selection (and thus is identical to <code>S1 and S2 around 12.3</code>) | ||
| + | |- | ||
| + | | <span style="color: #999">S1</span> beyond <span style="color: #999">12.3</span> of <span style="color: #999">S2</span> || be. | ||
| + | | Atoms in S1 that are at least 12.3 Anstroms away from S2 | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Properties | ||
| + | |- | ||
| + | | partial_charge <span style="color: #999">< 1.2</span> || pc. | ||
| + | | | ||
| + | |- | ||
| + | | formal_charge <span style="color: #999">= 1</span> || fc. | ||
| + | | | ||
| + | |- | ||
| + | | b <span style="color: #999">< 100.0</span> || | ||
| + | | B-factor less than 100.0 | ||
| + | |- | ||
| + | | q <span style="color: #999">< 1.0</span> || | ||
| + | | Occupancy less than 1.0 | ||
| + | |- | ||
| + | | ss <span style="color: #999">H+S</span> || | ||
| + | | Atoms with secondary structure H (helix) or S (sheet) | ||
| + | |- | ||
| + | | elem <span style="color: #999">C</span> || e. | ||
| + | | Atoms of element C (carbon) | ||
| + | |- | ||
| + | | p<span style="color: #999">.foo</span> = <span style="color: #999">12</span> || | ||
| + | | | ||
| + | |- | ||
| + | | p<span style="color: #999">.foo</span> < <span style="color: #999">12.3</span> || | ||
| + | | | ||
| + | |- | ||
| + | | p<span style="color: #999">.foo</span> in <span style="color: #999">12+34</span> || | ||
| + | | | ||
| + | |- | ||
| + | | stereo <span style="color: #999">R</span> || | ||
| + | | Chiral R/S stereo center with label R ''(only [https://pymol.org/d/media:stereochemistry Incentive PyMOL 1.4-1.8])'' | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Flags | ||
| + | |- | ||
| + | | bonded || | ||
| + | | Atoms which have at least one bond | ||
| + | |- | ||
| + | | protected || | ||
| + | | see [[protect]] | ||
| + | |- | ||
| + | | fixed || fxd. | ||
| + | | see [[flag]] | ||
| + | |- | ||
| + | | restrained || rst. | ||
| + | | see [[flag]] | ||
| + | |- | ||
| + | | masked || msk. | ||
| + | | see [[mask]] | ||
| + | |- | ||
| + | | flag <span style="color: #999">25</span> || f. | ||
| + | | Atoms with flag 25, see [[flag]] | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Chemical classes | ||
| + | |- | ||
| + | | organic || org. | ||
| + | | Non-polymer organic compounds (e.g. ligands, buffers) | ||
| + | |- | ||
| + | | inorganic || ino. | ||
| + | | Non-polymer inorganic atoms/ions | ||
| + | |- | ||
| + | | solvent || sol. | ||
| + | | Water molecules | ||
| + | |- | ||
| + | | polymer || pol. | ||
| + | | Protein or Nucleic Acid | ||
| + | |- | ||
| + | | polymer.protein || | ||
| + | | Protein ''(New in PyMOL 2.1)'' | ||
| + | |- | ||
| + | | polymer.nucleic || | ||
| + | | Nucleic Acid ''(New in PyMOL 2.1)'' | ||
| + | |- | ||
| + | | guide || | ||
| + | | Protein CA and nucleic acid C4*/C4' | ||
| + | |- | ||
| + | | hetatm || | ||
| + | | Atoms loaded from PDB HETATM records | ||
| + | |- | ||
| + | | hydrogens || h. | ||
| + | | Hydrogen atoms | ||
| + | |- | ||
| + | | backbone || bb. | ||
| + | | Polymer backbone atoms ''(new in PyMOL 1.6.1)'' | ||
| + | |- | ||
| + | | sidechain || sc. | ||
| + | | Polymer non-backbone atoms ''(new in PyMOL 1.6.1)'' | ||
| + | |- | ||
| + | | metals || | ||
| + | | Metal atoms ''(new in PyMOL 1.6.1)'' | ||
| + | |- | ||
| + | | donors || don. | ||
| + | | Hydrogen bond donor atoms | ||
| + | |- | ||
| + | | acceptors || acc. | ||
| + | | Hydrogen bond acceptor atoms | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Style | ||
| + | |- | ||
| + | | visible || v. | ||
| + | | Atoms in enabled objects with at least one visible representation | ||
| + | |- | ||
| + | | rep <span style="color: #999">cartoon</span> || | ||
| + | | Atoms with cartoon representation | ||
| + | |- | ||
| + | | color <span style="color: #999">blue</span> || | ||
| + | | Atoms with atom-color blue (by color index) | ||
| + | |- | ||
| + | | cartoon_color <span style="color: #999">blue</span> || | ||
| + | | Atoms with atom-level cartoon_color setting (by color index) | ||
| + | |- | ||
| + | | ribbon_color <span style="color: #999">blue</span> || | ||
| + | | Atoms with atom-level ribbon_color setting (by color index) | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Non molecular | ||
| + | |- | ||
| + | | center || | ||
| + | | Pseudo-atom at the center of the scene | ||
| + | |- | ||
| + | | origin || | ||
| + | | Pseudo-atom at the origin of rotation | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Coordinates | ||
| + | |- | ||
| + | | state <span style="color: #999">123</span> || | ||
| + | | Atoms with coordinates in state 123 | ||
| + | |- | ||
| + | | present || pr. | ||
| + | | Atoms with coordinates in the current state | ||
| + | |- | ||
| + | | x <span style="color: #999">< 12.3</span> || | ||
| + | | Atoms with model-space x coordinate less than 12.3 | ||
| + | |- | ||
| + | | y <span style="color: #999">< 12.3</span> || | ||
| + | | Atoms with model-space y coordinate less than 12.3 | ||
| + | |- | ||
| + | | z <span style="color: #999">> 12.3</span> || | ||
| + | | Atoms with model-space z coordinate greater than 12.3 | ||
| + | |- | ||
| + | !colspan="3" style="text-align:left"| Atom typing | ||
| + | |- | ||
| + | | text_type <span style="color: #999">TT</span> || tt. | ||
| + | | ''Auto-assigned in [https://pymol.org/d/media:atomtyping Incentive PyMOL 1.4-1.8])'' | ||
| + | |- | ||
| + | | numeric_type <span style="color: #999">123</span> || nt. | ||
| + | | | ||
| + | |} | ||
== Comparison of distance operators == | == Comparison of distance operators == | ||
| Line 124: | Line 341: | ||
| expand || ≤ X || center || always || 2 || | | expand || ≤ X || center || always || 2 || | ||
|} | |} | ||
| + | |||
| + | == Language Properties == | ||
| + | |||
| + | * names and keywords are case-insensitive unless [[ignore_case]] is set | ||
| + | * names and keywords can be abbreviated to non-ambiguous prefixes | ||
| + | |||
| + | '''Best practice recommendation:''' Only write case-sensitive, non-abbreviated selection expressions. That way your scripts will be robust against run-time configuration and future changes to the language (like addition of new keywords). | ||
== Examples == | == Examples == | ||
Logical selections can be combined. For example, you might select atoms that are part of chain a, but not residue number 125: | Logical selections can be combined. For example, you might select atoms that are part of chain a, but not residue number 125: | ||
<source lang="python"> | <source lang="python"> | ||
| − | # selects atoms that are part of chain | + | # selects atoms that are part of chain A, but not residue number 125. |
| − | select chain | + | select chain A and (not resi 125) |
# The following two selections are equivalent, | # The following two selections are equivalent, | ||
| − | select (name | + | select (name CB or name CG1 or name CG2) and chain A |
# select c-beta's, c-gamma-1's and c-gamma-2's | # select c-beta's, c-gamma-1's and c-gamma-2's | ||
# that are in chain A. | # that are in chain A. | ||
| − | select name | + | select name CB+CG1+CG2 and chain A |
# select all residues within 5 Ang. or any organic small molecules | # select all residues within 5 Ang. or any organic small molecules | ||
| Line 142: | Line 366: | ||
# select helices | # select helices | ||
| − | select ss ' | + | select ss 'H' |
# select anything shown as a line | # select anything shown as a line | ||
| Line 154: | Line 378: | ||
# select the 1st arginine | # select the 1st arginine | ||
| − | select first resn | + | select first resn ARG |
# select 1foo's segment G's chain X's residue 444's alpha carbon | # select 1foo's segment G's chain X's residue 444's alpha carbon | ||
| Line 170: | Line 394: | ||
Like the results of groups of arithmetic operations, the results of groups of logical operations depend on which operation is performed first. They have an order of precedence. To ensure that the operations are performed in the order you have in mind, use parentheses: | Like the results of groups of arithmetic operations, the results of groups of logical operations depend on which operation is performed first. They have an order of precedence. To ensure that the operations are performed in the order you have in mind, use parentheses: | ||
| − | <source lang="python">byres ((chain | + | <source lang="python">byres ((chain A or (chain B and (not resi 125))) around 5)</source> |
PyMOL will expand its logical selection out from the innermost parentheses. | PyMOL will expand its logical selection out from the innermost parentheses. | ||
| Line 176: | Line 400: | ||
== See Also == | == See Also == | ||
| − | * [[ | + | * [[select]] |
* [[Selection Macros]] | * [[Selection Macros]] | ||
* [[Property_Selectors]] | * [[Property_Selectors]] | ||
| + | * [[Selection Language Comparison]] with other modelling applications | ||
| + | * [[Identify]] | ||
[[Category:Selector Quick Reference]] | [[Category:Selector Quick Reference]] | ||
[[Category:Selecting|Selection Algebra]] | [[Category:Selecting|Selection Algebra]] | ||
Latest revision as of 03:15, 5 March 2018
PyMOL's selection language allows to select atoms based on identifiers and properties. Many commands (like color, show, etc.) take an atom selection argument to only operate on a subset of all atoms in the scene. Example:
PyMOL>show spheres, solvent and chain A
Selections can be made more precise or inclusive by combining them with logical operators, including the boolean and, or, and not. The boolean and selects only those items that have both (or all) of the named properties, and the boolean or selects items that have either (or any) of them.
Selection Operator/Modifier Table
Selection operators and modifiers are listed below. The dummy variables s1 and s2 stand for selection-expressions such as "chain a" or "hydro."
| Operator | Aliases | Description |
|---|---|---|
| Generic | ||
| all | * | All atoms currently loaded into PyMOL |
| none | Empty selection | |
| enabled | Atoms from enabled objects | |
| Named selections | ||
| sele | Named selection or object "sele", but only if it doesn't conflict with the name of another operator | |
| %sele | Named selection or object "sele" Recommended, avoids ambiguity | |
| ?sele | Named selection or object "sele", or empty selection if "sele" doesn't exist | |
| Logical | ||
| not S1 | ! | Inverts selection |
| S1 and S2 | & | Atoms included in both S1 and S2 |
| S1 or S2 | | | Atoms included in either S1 or S2 |
| S1 S2 | implicit or | |
| S1 and (S2 or S3) | Parentheses for evaluation order control | |
| first S1 | First atom in S1 (single atom selection) | |
| last S1 | Last atom in S1 (single atom selection) | |
| Identifiers (see also Selection Macros) | ||
| model 1ubq | m. | Atoms from object "1ubq" |
| chain C | c. | Chain identifier "C" |
| segi S | s. | Segment identifier "S" (label_asym_id from mmCIF) |
| resn ALA | r. | Residue name "ALA" |
| resi 100-200 | i. | Residue identifier between 100 and 200 |
| name CA | n. | Atom name "CA" |
| alt A | Alternate location "A" | |
| index 123 | idx. | Internal per-object atom index (changes with sorting) |
| id 123 | ID column from PDB file | |
| rank 123 | Per-object atom index at load time (see also retain_order) | |
| pepseq ACDEF | ps. | Protein residue sequence with one-letter code "ACDEF" (see also FindSeq) |
| label "Hello World" | Atoms with label "Hello World" (new in PyMOL 1.9) | |
| Identifier matching | ||
| S1 in S2 | Atoms in S1 whose identifiers name, resi, resn, chain and segi all match atoms in S2 | |
| S1 like S2 | Atoms in S1 whose identifiers name and resi match atoms in S2 | |
| Entity expansion | ||
| Important: All "by"-operators have a weak priority, so (byres S1 or S2) is actually identical to (byres (S1 or S2)) and not to ((byres S1) or S2) | ||
| byobject S1 | Expands S1 to complete objects | |
| bysegi S1 | bs. | Expands S1 to complete segments |
| bychain S1 | bc. | Expands S1 to complete chains |
| byres S1 | br. | Expands S1 to complete residues |
| bycalpha S1 | bca. | CA atoms of residues with at least one atom in S1 |
| bymolecule S1 | bm. | Expands S1 to complete molecules (connected with bonds) |
| byfragment S1 | bf. | |
| byring S1 | All rings of size ≤ 7 which have at least one atom in S1 (new in PyMOL 1.8.2) | |
| bycell S1 | Expands selection to unit cell | |
| Bond expansion | ||
| bound_to S1 | bto. | Atoms directly bonded to S1, may include S1 |
| neighbor S1 | nbr. | Atoms directly bonded to S1, excludes S1 |
| S1 extend 3 | xt. | Expands S1 by 3 bonds connected to atoms in S1 |
| Proximity (see also comparison of distance operators) | ||
| S1 within 12.3 of S2 | w. | Atoms in S1 that are within 12.3 Angstroms of any atom in S2 |
| S1 around 12.3 | a. | Atoms with centers within 12.3 Angstroms of the center of any atom in S1 |
| S1 expand 12.3 | x. | Expands S1 by atoms within 12.3 Angstroms of the center of any atom in S1 |
| S1 gap 1.2 | Atoms whose VDW radii are separated from the VDW radii of S1 by a minimum of 1.2 Angstroms. | |
| S1 near_to 12.3 of S2 | nto. | Same as within, but excludes S2 from the selection (and thus is identical to S1 and S2 around 12.3)
|
| S1 beyond 12.3 of S2 | be. | Atoms in S1 that are at least 12.3 Anstroms away from S2 |
| Properties | ||
| partial_charge < 1.2 | pc. | |
| formal_charge = 1 | fc. | |
| b < 100.0 | B-factor less than 100.0 | |
| q < 1.0 | Occupancy less than 1.0 | |
| ss H+S | Atoms with secondary structure H (helix) or S (sheet) | |
| elem C | e. | Atoms of element C (carbon) |
| p.foo = 12 | ||
| p.foo < 12.3 | ||
| p.foo in 12+34 | ||
| stereo R | Chiral R/S stereo center with label R (only Incentive PyMOL 1.4-1.8) | |
| Flags | ||
| bonded | Atoms which have at least one bond | |
| protected | see protect | |
| fixed | fxd. | see flag |
| restrained | rst. | see flag |
| masked | msk. | see mask |
| flag 25 | f. | Atoms with flag 25, see flag |
| Chemical classes | ||
| organic | org. | Non-polymer organic compounds (e.g. ligands, buffers) |
| inorganic | ino. | Non-polymer inorganic atoms/ions |
| solvent | sol. | Water molecules |
| polymer | pol. | Protein or Nucleic Acid |
| polymer.protein | Protein (New in PyMOL 2.1) | |
| polymer.nucleic | Nucleic Acid (New in PyMOL 2.1) | |
| guide | Protein CA and nucleic acid C4*/C4' | |
| hetatm | Atoms loaded from PDB HETATM records | |
| hydrogens | h. | Hydrogen atoms |
| backbone | bb. | Polymer backbone atoms (new in PyMOL 1.6.1) |
| sidechain | sc. | Polymer non-backbone atoms (new in PyMOL 1.6.1) |
| metals | Metal atoms (new in PyMOL 1.6.1) | |
| donors | don. | Hydrogen bond donor atoms |
| acceptors | acc. | Hydrogen bond acceptor atoms |
| Style | ||
| visible | v. | Atoms in enabled objects with at least one visible representation |
| rep cartoon | Atoms with cartoon representation | |
| color blue | Atoms with atom-color blue (by color index) | |
| cartoon_color blue | Atoms with atom-level cartoon_color setting (by color index) | |
| ribbon_color blue | Atoms with atom-level ribbon_color setting (by color index) | |
| Non molecular | ||
| center | Pseudo-atom at the center of the scene | |
| origin | Pseudo-atom at the origin of rotation | |
| Coordinates | ||
| state 123 | Atoms with coordinates in state 123 | |
| present | pr. | Atoms with coordinates in the current state |
| x < 12.3 | Atoms with model-space x coordinate less than 12.3 | |
| y < 12.3 | Atoms with model-space y coordinate less than 12.3 | |
| z > 12.3 | Atoms with model-space z coordinate greater than 12.3 | |
| Atom typing | ||
| text_type TT | tt. | Auto-assigned in Incentive PyMOL 1.4-1.8) |
| numeric_type 123 | nt. | |
Comparison of distance operators
There are serveral very similar operators that select by pairwise atom distances. The following table lists the details how they differ.
Syntax 1: s1 operator X of s2
Syntax 2: s1 and (s2 operator X)
| operator | distance is ... | measured from | includes s2 | syntax | notes |
|---|---|---|---|---|---|
| near_to | ≤ X | center | never | 1 | equivalent to "around" |
| within | ≤ X | center | if matches s1 | 1 | |
| beyond | > X | center | never | 1 | |
| gap | > X | center+vdw | never | 2 | |
| around | ≤ X | center | never | 2 | equivalent to "near_to" |
| expand | ≤ X | center | always | 2 |
Language Properties
- names and keywords are case-insensitive unless ignore_case is set
- names and keywords can be abbreviated to non-ambiguous prefixes
Best practice recommendation: Only write case-sensitive, non-abbreviated selection expressions. That way your scripts will be robust against run-time configuration and future changes to the language (like addition of new keywords).
Examples
Logical selections can be combined. For example, you might select atoms that are part of chain a, but not residue number 125:
# selects atoms that are part of chain A, but not residue number 125.
select chain A and (not resi 125)
# The following two selections are equivalent,
select (name CB or name CG1 or name CG2) and chain A
# select c-beta's, c-gamma-1's and c-gamma-2's
# that are in chain A.
select name CB+CG1+CG2 and chain A
# select all residues within 5 Ang. or any organic small molecules
select br. all within 5 of organic
# select helices
select ss 'H'
# select anything shown as a line
select rep lines
# select all residues with a b-factor less than 20, within 3 angstroms of any water
select br. b<20 & (all within 3 of resn HOH)
# select anything colored blue
select color blue
# select the 1st arginine
select first resn ARG
# select 1foo's segment G's chain X's residue 444's alpha carbon
select 1foo/G/X/444/CA
# same thing
select 1foo and segi G and c. X and i. 444 and n. CA
# select the entire object that residue 23's beta caron is in:
select bo. i. 23 and n. CA
# select the molecule that chain C is in
select bm. c. C
Like the results of groups of arithmetic operations, the results of groups of logical operations depend on which operation is performed first. They have an order of precedence. To ensure that the operations are performed in the order you have in mind, use parentheses:
byres ((chain A or (chain B and (not resi 125))) around 5)
PyMOL will expand its logical selection out from the innermost parentheses.
See Also
- select
- Selection Macros
- Property_Selectors
- Selection Language Comparison with other modelling applications
- Identify