This is a read-only mirror of pymolwiki.org
Difference between revisions of "Ray"
(notes about performance and distance) |
m (2 revisions) |
||
| (26 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | '''ray''' creates a ray-traced image of the current frame. | |
| − | '''ray''' creates a ray-traced image of the current frame. This command is used to make high-resolution photos fit for publication and formal movies. Please note, the '''ray''' command can take some time (up to several minutes, depending on image complexity and size). | + | [[Image:Gslike.png|right|350px|thumb|Varying settings to play with rendering options]] |
| + | |||
| + | =Details= | ||
| + | This command is used to make high-resolution photos fit for publication and formal movies. Please note, the '''ray''' command can take some time (up to several minutes, depending on image complexity and size). | ||
For those who are making movies with PyMOL, '''Ray''' is one of the most commonly used last steps before stitching the frames together to compile the movie. '''Ray''' has many powerful features such as setting the size of the image -- and it still works even if the [[Viewport]] or screen is smaller than the requested output file dimensions. | For those who are making movies with PyMOL, '''Ray''' is one of the most commonly used last steps before stitching the frames together to compile the movie. '''Ray''' has many powerful features such as setting the size of the image -- and it still works even if the [[Viewport]] or screen is smaller than the requested output file dimensions. | ||
| − | == | + | [[Image:No_ray_trace.png|Image, not ray traced.|thumb|200px]] [[Image:ray_traced.png|Image, ray traced.|thumb|200px]] |
| + | |||
| + | ==Usage== | ||
ray [width,height [,renderer [,angle [,shift ]]] | ray [width,height [,renderer [,angle [,shift ]]] | ||
'''angle''' and '''shift''' can be used to generate matched stereo pairs | '''angle''' and '''shift''' can be used to generate matched stereo pairs | ||
| + | '''width''' and '''height''' can be set to any non-negative integer. If both are set to zero than the current window size is used and is equivalent to just using '''ray''' with no arguments. If one is set to zero (or missing) while the other is a positive integer, then the argument set to zero (or missing) will be scaled to preserve the current aspect ratio. | ||
| − | == | + | ==PyMol API== |
<source lang="python">cmd.ray(int width,int height,int renderer=-1,float shift=0)</source> | <source lang="python">cmd.ray(int width,int height,int renderer=-1,float shift=0)</source> | ||
| − | == | + | ==Settings== |
===Modes=== | ===Modes=== | ||
Setting the '''[[Ray_trace_mode]]''' variable in PyMOL changes the way PyMOL's internal renderer represents proteins in the final output. New modes were recently added to give the user more options of molecular representation. New modes are: normal rendering, but with a black outline (nice for presentations); black and white only; quantized color with black outline (also, very nice for presentations; more of a ''cartoony'' appearance). | Setting the '''[[Ray_trace_mode]]''' variable in PyMOL changes the way PyMOL's internal renderer represents proteins in the final output. New modes were recently added to give the user more options of molecular representation. New modes are: normal rendering, but with a black outline (nice for presentations); black and white only; quantized color with black outline (also, very nice for presentations; more of a ''cartoony'' appearance). | ||
| Line 33: | Line 39: | ||
set ray_trace_mode, 1 # (or 2 or 3; best with "bg_color white;set antialias,2") | set ray_trace_mode, 1 # (or 2 or 3; best with "bg_color white;set antialias,2") | ||
# These two new modes -- 2 and 3 -- are cool cartoon looking modes. | # These two new modes -- 2 and 3 -- are cool cartoon looking modes. | ||
| + | |||
| + | # change the color of the outline to a named color, or a hex-code | ||
| + | set ray_trace_color, magenta | ||
| + | set ray_trace_color, 0x0033ff | ||
</source> | </source> | ||
| Line 66: | Line 76: | ||
'''renderer = 0''' uses PyMOL's internal renderer | '''renderer = 0''' uses PyMOL's internal renderer | ||
| − | '''renderer = 1''' uses PovRay's renderer. This is Unix-only and you must have " | + | '''renderer = 1''' uses PovRay's renderer. This is Unix-only and you must have "povray" in your path. It utilizes two temporary files: "tmp_pymol.pov" and "tmp_pymol.png". Also works on Mac via Povray37UnofficialMacCmd but it needs to be in your path as "povray". |
==Performance== | ==Performance== | ||
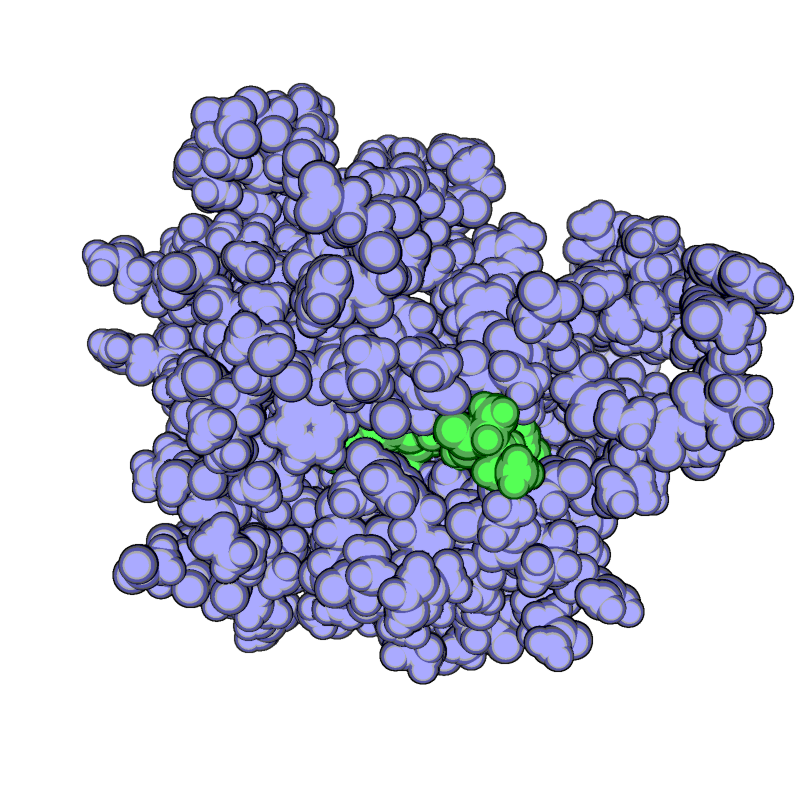
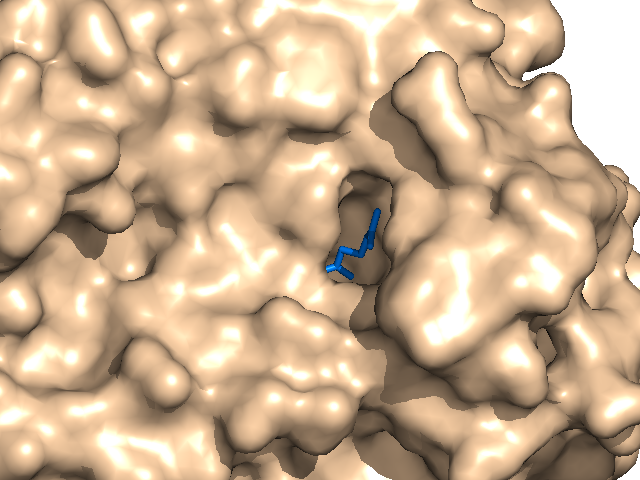
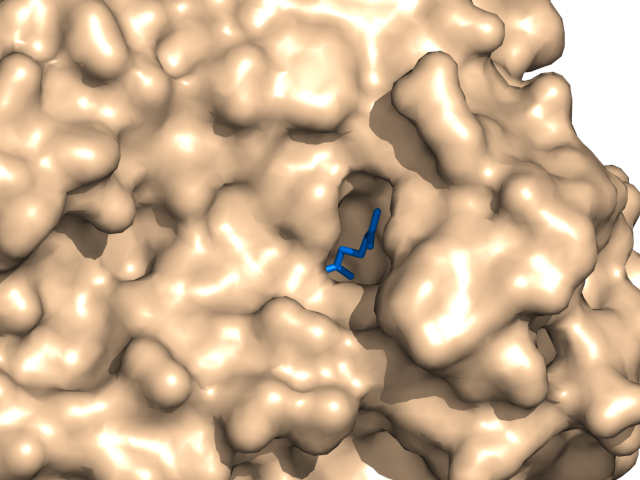
| − | The ray performance depends on distance between camera and molecule. | + | *The ray performance depends on distance between camera and molecule. |
If the distance is big rendering takes much time. If the distance is too small distant parts of molecule dissolve. | If the distance is big rendering takes much time. If the distance is too small distant parts of molecule dissolve. | ||
| − | |||
<gallery> | <gallery> | ||
Image:Close_ray.png|Too close to molecule | Image:Close_ray.png|Too close to molecule | ||
Image:Middle_ray.png|Normal distance | Image:Middle_ray.png|Normal distance | ||
</gallery> | </gallery> | ||
| + | * Tip: If you have a rather complicated scene that is zoomed into only a part of the molecule, you can speed up the ray tracing by hiding everything else outside of a certain range of the zoomed-on point. For example, if I have a large molecule and I'm looking only at the 30-atom ligand bound to it, then I can do something like the following: | ||
| + | <source lang="python"> | ||
| + | # setup your complex scene | ||
| + | ... | ||
| + | |||
| + | # zoom on the hetero atom (ligand and not water) within 5 Angstroms | ||
| + | select hh, het and not resn HOH | ||
| + | zoom hh, 5 | ||
| + | |||
| + | # turn on depth cueing | ||
| + | set depth_cue, 1 | ||
| + | |||
| + | # now, select stuff to hide; we select everything that is | ||
| + | # farther than 8 Ang from our main selection | ||
| + | select th, (all) and not ( (all) within 8 of hh) ) | ||
| + | |||
| + | hide everything, th | ||
| + | |||
| + | # any additional commands you want | ||
| + | ... | ||
| + | |||
| + | ray | ||
| + | </source> | ||
| + | As an example of the efficacy of this method, I ray traced a rather complex scene with all the atoms visible here's the output of ray: | ||
| + | <source lang="bash"> | ||
| + | PyMOL>ray | ||
| + | Ray: render time: 24.50 sec. = 146.9 frames/hour (941.88 sec. accum.). | ||
| + | </source> | ||
| + | and here is the result when I soft-clipped everything else using the above method: | ||
| + | <source lang="bash"> | ||
| + | PyMOL>ray | ||
| + | Ray: render time: 47.93 sec. = 75.1 frames/hour (989.80 sec. accum.). | ||
| + | </source> | ||
| + | The two images in the following gallery show the results of the rendering. | ||
| + | <gallery> | ||
| + | Image:Ray_method_off.png|normal ray tracing. This took twice as long to make as the image to the right. Same size, and DPI. | ||
| + | Image:Ray_method_on.png|manually hiding things you won't see anyway. This took 1/2 the time to render as compared to the same sized & DPId image at left. | ||
| + | </gallery> | ||
| + | |||
| + | ===Memory=== | ||
| + | If memory is an issue for you in PyMOL, try executing your rendering from a script rather than a PyMOL session file. An unfortunate unavoidable consequence of the fact that we use Python's portable, platform-independent "pickle" machinery for PyMOL session files. Packing or unpacking a Session or Scene file thus requires that there be two simultanous copies of the information to reside in RAM simultaneously: one native and a second in Python itself. | ||
| + | |||
| + | So when memory is a limiting factor, scripts are recommended over sessions. | ||
==Examples== | ==Examples== | ||
| Line 126: | Line 178: | ||
===Ray Tracing Stereo Images=== | ===Ray Tracing Stereo Images=== | ||
| − | See [[Stereo_Ray]] | + | :''See [[Stereo_Ray]]'' |
| − | |||
| − | |||
| − | |||
| − | |||
| − | See also [[Ray Tracing]]. | + | ==See also== |
| + | # "help faster" for optimization tips with the builtin renderer. "help povray" for how to use PovRay instead of PyMOL's built-in ray-tracing engine. For high-quality photos, please also see the [[Antialias]] command. [[Ray shadows]] for controlling shadows. | ||
| + | # See also [[Ray Tracing]]. | ||
| + | # [http://www.gimp.org/tutorials/Color2BW Desaturation Tutorial] -- A good resource for making nice B&W images from color images (desaturation). | ||
| + | #[[Ray Trace Gain]] | ||
| − | == | + | ==User comments== |
| − | + | ;How do I ray trace a publication-ready (~300dpi) image using PyMol? | |
| − | This answer is in the [[:Category:Advanced_Issues|Advanced Issues]] (Image Manipulation Section). | + | :This answer is in the [[:Category:Advanced_Issues|Advanced Issues]] (Image Manipulation Section). |
| − | [[Category:Commands| | + | [[Category:Commands|Ray]] |
| − | [[Category:Publication_Quality| | + | [[Category:Publication_Quality|Ray]] |
| + | [[Category:Performance|Ray]] | ||
| + | [[Category:Movies|Ray]] | ||
Latest revision as of 15:32, 20 October 2014
ray creates a ray-traced image of the current frame.
Details
This command is used to make high-resolution photos fit for publication and formal movies. Please note, the ray command can take some time (up to several minutes, depending on image complexity and size).
For those who are making movies with PyMOL, Ray is one of the most commonly used last steps before stitching the frames together to compile the movie. Ray has many powerful features such as setting the size of the image -- and it still works even if the Viewport or screen is smaller than the requested output file dimensions.
Usage
ray [width,height [,renderer [,angle [,shift ]]]
angle and shift can be used to generate matched stereo pairs
width and height can be set to any non-negative integer. If both are set to zero than the current window size is used and is equivalent to just using ray with no arguments. If one is set to zero (or missing) while the other is a positive integer, then the argument set to zero (or missing) will be scaled to preserve the current aspect ratio.
PyMol API
cmd.ray(int width,int height,int renderer=-1,float shift=0)
Settings
Modes
Setting the Ray_trace_mode variable in PyMOL changes the way PyMOL's internal renderer represents proteins in the final output. New modes were recently added to give the user more options of molecular representation. New modes are: normal rendering, but with a black outline (nice for presentations); black and white only; quantized color with black outline (also, very nice for presentations; more of a cartoony appearance).
Note: Mode 3, the quantized color one, sort of burns the background if you're using this setting. This will make a pure white background somewhat "offwhite"; thus, a poster would look poor because you could see the border for the image. If you'll be using this mode, try the ray_opaque_background setting.
# normal color
set ray_trace_mode, 0
# normal color + black outline
set ray_trace_mode, 1
# black outline only
set ray_trace_mode, 2
# quantized color + black outline
set ray_trace_mode, 3
set ray_trace_mode, 1 # (or 2 or 3; best with "bg_color white;set antialias,2")
# These two new modes -- 2 and 3 -- are cool cartoon looking modes.
# change the color of the outline to a named color, or a hex-code
set ray_trace_color, magenta
set ray_trace_color, 0x0033ff
Here are the example images for the new modes
Perspective
Perspective Example Images
Notes
PyMol 0.97 and prior used orthoscopic rendering -- that is, no perspective. Upon the arrival of 0.98 and later, we get perspective based rendering at a cost of a 4x decrease in render speed. If you want perspective
set orthoscopic, off
Otherwise
set orthoscopic, on
To magnify the effect of perspective on the scene,
set field_of_view, X
where 50<X<70. Default is 20. 50-70 gives a very strong perspective effect. Nb. the field of view is in Y, not X as one would expect.
Renderer
renderer = -1 is default (use value in ray_default_renderer)
renderer = 0 uses PyMOL's internal renderer
renderer = 1 uses PovRay's renderer. This is Unix-only and you must have "povray" in your path. It utilizes two temporary files: "tmp_pymol.pov" and "tmp_pymol.png". Also works on Mac via Povray37UnofficialMacCmd but it needs to be in your path as "povray".
Performance
- The ray performance depends on distance between camera and molecule.
If the distance is big rendering takes much time. If the distance is too small distant parts of molecule dissolve.
- Tip: If you have a rather complicated scene that is zoomed into only a part of the molecule, you can speed up the ray tracing by hiding everything else outside of a certain range of the zoomed-on point. For example, if I have a large molecule and I'm looking only at the 30-atom ligand bound to it, then I can do something like the following:
# setup your complex scene
...
# zoom on the hetero atom (ligand and not water) within 5 Angstroms
select hh, het and not resn HOH
zoom hh, 5
# turn on depth cueing
set depth_cue, 1
# now, select stuff to hide; we select everything that is
# farther than 8 Ang from our main selection
select th, (all) and not ( (all) within 8 of hh) )
hide everything, th
# any additional commands you want
...
ray
As an example of the efficacy of this method, I ray traced a rather complex scene with all the atoms visible here's the output of ray:
PyMOL>ray
Ray: render time: 24.50 sec. = 146.9 frames/hour (941.88 sec. accum.).
and here is the result when I soft-clipped everything else using the above method:
PyMOL>ray
Ray: render time: 47.93 sec. = 75.1 frames/hour (989.80 sec. accum.).
The two images in the following gallery show the results of the rendering.
Memory
If memory is an issue for you in PyMOL, try executing your rendering from a script rather than a PyMOL session file. An unfortunate unavoidable consequence of the fact that we use Python's portable, platform-independent "pickle" machinery for PyMOL session files. Packing or unpacking a Session or Scene file thus requires that there be two simultanous copies of the information to reside in RAM simultaneously: one native and a second in Python itself.
So when memory is a limiting factor, scripts are recommended over sessions.
Examples
Simple
# ray trace the current scene using the default size of the viewport
ray
Specify Image Size
# ray trace the current scene, but scaled to 1024x768 pixels
ray 1024,768
Specify Renderer
# ray trace with an external renderer.
ray renderer=0
High Quality B&W Rendering
# Black and White Script
load /tmp/3fib.pdb;
show cartoon;
set ray_trace_mode, 2; # black and white cartoon
bg_color white;
set antialias, 2;
ray 600,600
png /tmp/1l9l.png
High Quality Color
# Color Script
load /tmp/thy_model/1l9l.pdb;
hide lines;
show cartoon;
set ray_trace_mode, 3; # color
bg_color white;
set antialias, 2;
remove resn HOH
remove resn HET
ray 600,600
png /tmp/1l9l.png
Ray Tracing Stereo Images
- See Stereo_Ray
See also
- "help faster" for optimization tips with the builtin renderer. "help povray" for how to use PovRay instead of PyMOL's built-in ray-tracing engine. For high-quality photos, please also see the Antialias command. Ray shadows for controlling shadows.
- See also Ray Tracing.
- Desaturation Tutorial -- A good resource for making nice B&W images from color images (desaturation).
- Ray Trace Gain
User comments
- How do I ray trace a publication-ready (~300dpi) image using PyMol?
- This answer is in the Advanced Issues (Image Manipulation Section).