This is a read-only mirror of pymolwiki.org
Difference between revisions of "PyMOLProbity"
Jaredsampson (talk | contribs) (Created page.) |
Jaredsampson (talk | contribs) (Add screenshot.) |
||
| Line 7: | Line 7: | ||
== Description == | == Description == | ||
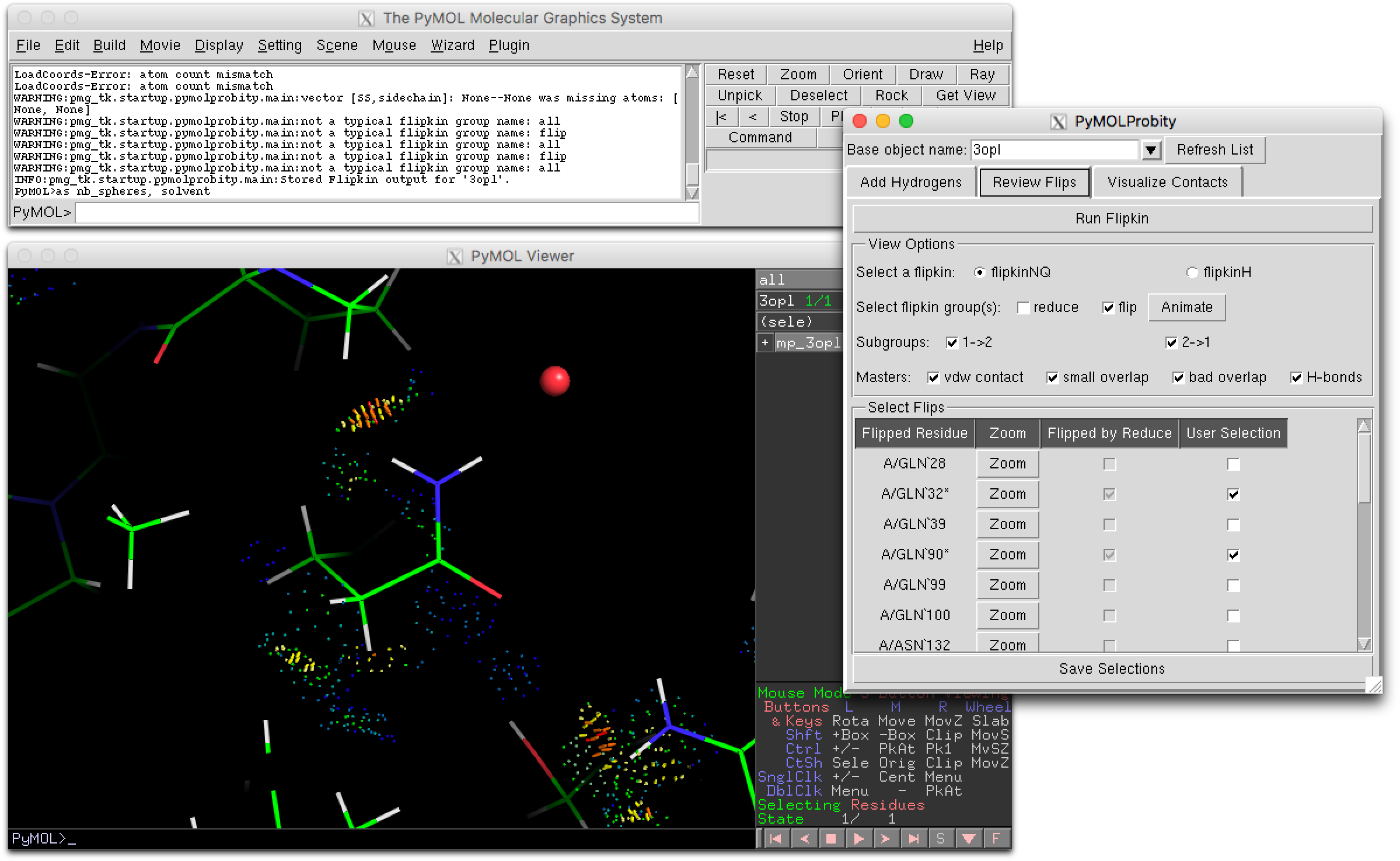
| − | [[ | + | [[File:PyMOLProbity_GUI.png|480px|thumb|right|The PyMOLProbity GUI can be used to inspect and adjust clashes and flip orientations of flippable side chain groups.]] |
PyMOLProbity is a plugin allows the user to produce MolProbity-style | PyMOLProbity is a plugin allows the user to produce MolProbity-style | ||
Revision as of 17:41, 14 January 2017
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/jaredsampson/pymolprobity/raw/master/pymolprobity.tar.gz |
| Author(s) | Jared Sampson |
| License | MIT |
Description
PyMOLProbity is a plugin allows the user to produce MolProbity-style visualization of atomic interactions within a structure (e.g. H-bonds, van der Waals interactions and clashes) directly within a PyMOL session. The plugin runs local copies of several executable programs from the Richardson Lab at Duke University, authors of the MolProbity software, parses the output, and displays the results in the PyMOL viewport. There are both a graphical user interface (GUI) for general point-and-click use, and a command-line interface (CLI) suitable for scripting.
Installation
For installation instructions, please see the README file in the repository.
Getting Started
First run
Now you can open PyMOL, load or fetch a structure, and launch PyMOLProbity from the Plugin menu.
- Use the Add Hydrogens tab to add hydrogens with Reduce. This will also calculate which N/Q/H residue side chains should be flipped.
- If you would like to examine these more closely, use the Review Flips tab to examine each flippable residue and choose the ones you wish to flip or keep.
- Save these using the Save Selections button.
- Finally, use the Visualize Contacts tab to run Probe on the modified coordinates and generate contact dots and clash vectors for all the atoms in your object.
Command-Line Interface
The plugin makes the following functions available:
- reduce_obj(obj, flip=1): Run reduce on a loaded PyMOL object (or named selection) with (default) or without making the Asn/Gln/His flips recommended by Reduce.
- flipkin_obj(obj): Run Flipkin to create both NQ and H flipkin kinemage visualization of the Reduce-modified structure.
- probe_obj(obj): Run Probe on either a structure saved from the Flipkin tab of the GUI, or the Reduce-modified structure.
Note that both `flipkin_obj` and `probe_obj` require previously having run `reduce_obj` on the same object.