This is a read-only mirror of pymolwiki.org
Difference between revisions of "Optimize"
m (→Change log) |
m (add citation information (to the openbabel paper)) |
||
| Line 37: | Line 37: | ||
*2013-10-24 (Version 0.2) | *2013-10-24 (Version 0.2) | ||
*# now openbabel add the hydrogen and not PyMOL. | *# now openbabel add the hydrogen and not PyMOL. | ||
| + | |||
| + | |||
| + | == Citation == | ||
| + | If you find optimize useful please consider citing this work | ||
| + | Noel M. O'Boyle , Michael Banck , Craig A. James , Chris Morley , Tim Vandermeersch and Geoffrey R. Hutchison. "Open Babel: An open chemical toolbox." Journal of Cheminformatics 2011, 3:33. http://www.jcheminf.com/content/3/1/33 | ||
| + | |||
[[Category:Plugins]] | [[Category:Plugins]] | ||
[[Category:Pymol-script-repo]] | [[Category:Pymol-script-repo]] | ||
Revision as of 22:15, 24 January 2014
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/optimize.py |
| Author(s) | Osvaldo Martin |
| License | GPL |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
Optimize provides a PyMOL graphical interface to some of the molecular mechanics features available in openbabel, allowing the user to minimize the energy of any molecule uploaded on PyMOL.
Installation
Optimize was tested only on GNU/Linux but should work on other operating systems. The plugin can be downloaded through the project Pymol-script-repo. Optimize needs openbabel to be installed on your computer (see instructions here).
Usage
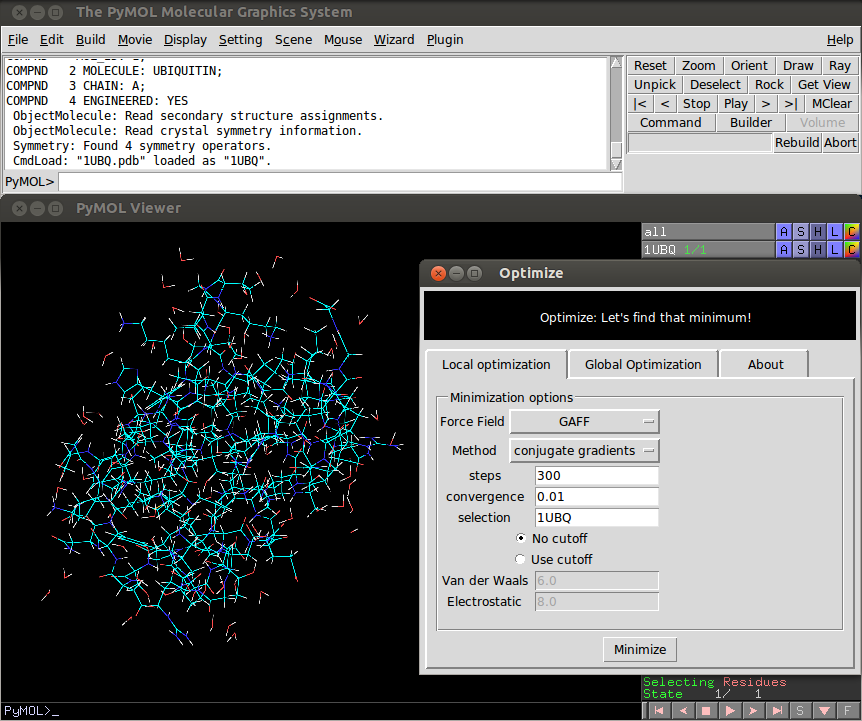
The plugin can be accessed using a graphic user interface (see figure 0) or from the PyMOL`s terminal trough the following command:
minimize [selection string [, forcefield string [, method string [, nsteps int [, conv float [, cutoff bool [, cut_vdw float [, cut_elec float]]]]]]]
Where:
- selection: Select what is going to be minimized. The default value is 'all'. Using the GUI the default value is the first uploaded object.
- forcefield: Select the forcefield used to compute the Internal Energy, options available are GAFF, MMFF94s, MMFF94, UFF and Ghemical.
- method: Select the method used to find the local minimum. The methods available are conjugate gradients and steepest descent.
- nsteps: Number of iteration steps during the minimization.
- conv: Criteria used to judge minimization convergence.
- cutoff: Control if cut-off are used or not to compute non-bonded interactions, posible values are True of False.
- cut_vdw: If cutoff is True, then this parameter set the limit beyond which two atoms do not interact trough Van der Waals forces.
- cut_elec: If cutoff is True, then this parameter set the limit beyond which two atoms do not interact trough electrostatic forces forces.
Change log
- 2013-10-06 (Version 0.1)
- First version. (More features coming soon!)
- 2013-10-24 (Version 0.2)
- now openbabel add the hydrogen and not PyMOL.
Citation
If you find optimize useful please consider citing this work Noel M. O'Boyle , Michael Banck , Craig A. James , Chris Morley , Tim Vandermeersch and Geoffrey R. Hutchison. "Open Babel: An open chemical toolbox." Journal of Cheminformatics 2011, 3:33. http://www.jcheminf.com/content/3/1/33