|
|
| (59 intermediate revisions by 4 users not shown) |
| Line 1: |
Line 1: |
| | + | {{Infobox script-repo |
| | + | |type = plugin |
| | + | |filename = plugins/mtsslWizard.py |
| | + | |author = [[User:Gha|Gregor Hagelueken]] |
| | + | |license = - |
| | + | }} |
| | = mtsslWizard = | | = mtsslWizard = |
| | mtsslWizard is a PyMOL wizard for ''in silico'' spin labeling of proteins. | | mtsslWizard is a PyMOL wizard for ''in silico'' spin labeling of proteins. |
| | | | |
| | ==News== | | ==News== |
| − | 2012-12-20:
| + | 2020-05-21 |
| | + | *If you are having trouble with installation give [http://www.mtsslsuite.isb.ukbonn.de this] a try |
| | | | |
| − | We are just working on Version 1.1 of mtsslWizard. Among other things, it will be much, much faster than version 1.0. It should come out in January or early February 2013.
| + | 2017-01-17: |
| | + | *[http://www-hagelueken.thch.uni-bonn.de/index.php/en/2014-08-04-13-28-34/mtssldock Version 2.0 available]! |
| | | | |
| − | ==Program features==
| + | 2015-07-27: |
| − | An MTSSL spin label can be attached to any position of a protein just by pointing and clicking. The program can then search the conformational space of the label (5 chi angles) and determine which conformations of the label do not clash with the protein surface. In "distance mode", distances between MTSSL ensembles can be determined and exported.
| |
| − | For symmetric molecules, the MTSSL ensemble can be copied to symmetry related positions simply by pointing and clicking ("Copy & Move" mode).
| |
| | | | |
| − | Please reference: [http://www.springerlink.com/content/7148j5k2ppk475wu/ '''MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL''', Gregor Hagelueken, Richard Ward, James H. Naismith and Olav Schiemann, DOI: 10.1007/s00723-012-0314-0]
| + | *uploaded a new version (1.3) to repository. Includes some new spin labels and a distance map feature. Plots can be viewed with [[mtsslPlotter]]. |
| | | | |
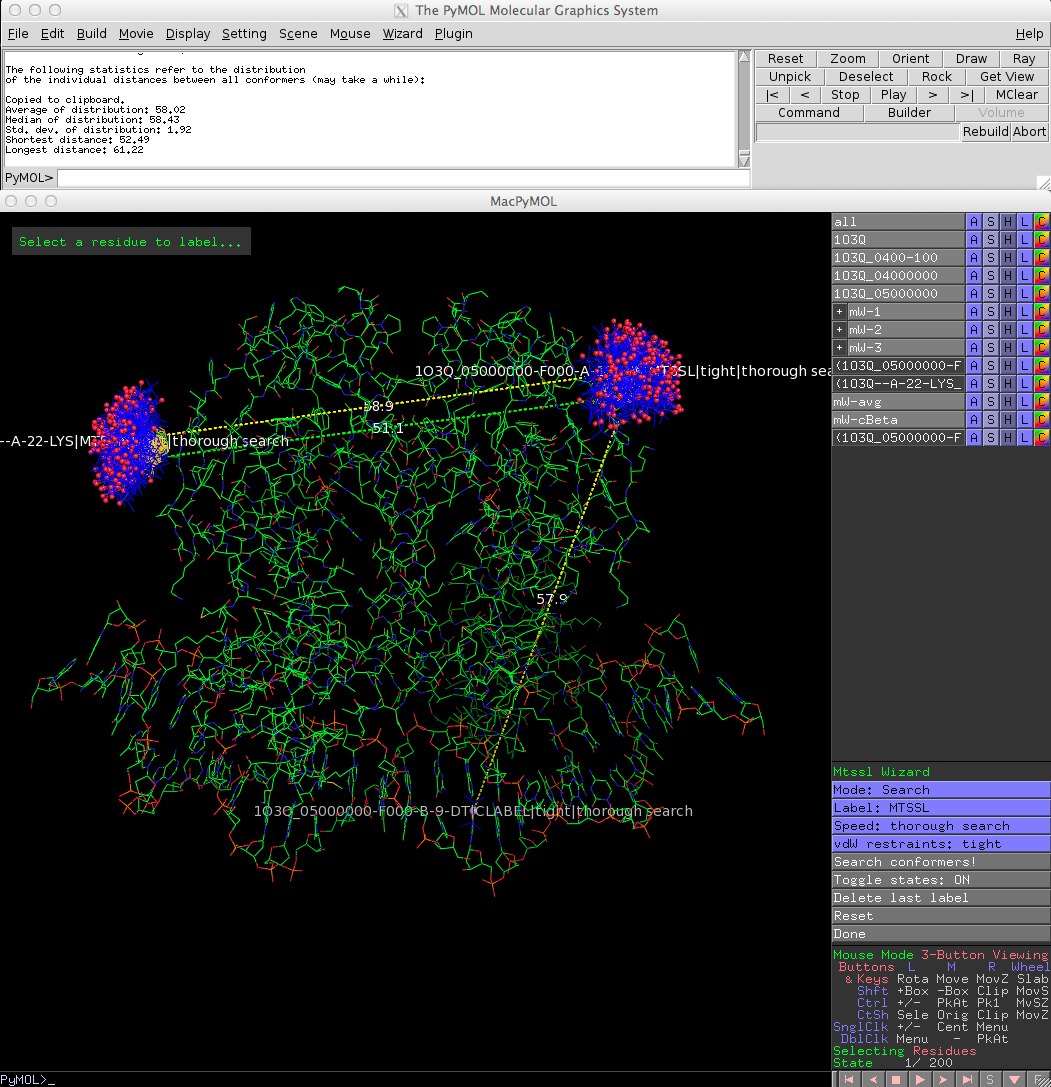
| − | ==Screen shot==
| + | 2014-03-04: |
| − | [[File:MtsslWizard_gui.jpg]]
| |
| | | | |
| − | ==Usage==
| + | * uploaded a new version (1.12) to repository. Fixes a bug that occurred on some machines. Thanks to C. Engelhard for noticing! |
| − | ===Installation===
| |
| − | Install the program by copying the code below into an empty text file (e.g. "mtsslWizard.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. mtsslWizard has been tested with PyMOL 1.4.
| |
| − | ===Labeling===
| |
| − | #Open a protein structure in PyMOL and remove any unwanted solvent molecules or ligands.
| |
| − | #Start the mtsslWizard via Wizards>mtsslWizard
| |
| − | #If needed, vary the search parameters like "Thoroughness", "Cutoff" and "Clashes allowed". Thoroughness determines how many conformations of MTSSL will be checked. The cutoff value determines the minimum allowed distance between label and protein before the conformation is sorted out as clashing. "Clashes allowed" should usually be set to zero. If this value is changed to e.g. "5", a conformation can violate the "cutoff" parameter five times before the conformation is discarded.
| |
| − | #click "Go" to start the calculation. Depending on the size of the protein, the "Thoroughness" parameter and of course the speed of your computer the duration of the search can vary quite considerably. With the default settings it does usually not take longer than 10-15 seconds.
| |
| − | ===Distance calculation===
| |
| − | #change "Mode" to "Distance"
| |
| − | #click the first and second ensemble
| |
| − | #The program will now calculate all possible distances between the conformations in the two ensembles. If both ensembles have many conformations, this can take quite a while!
| |
| − | ===Reference===
| |
| − | If you find this program useful, please cite this paper:
| |
| | | | |
| − | Hagelueken G, Ward R, Naismith JH, Schiemann O (2012) MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. Appl. Magn. Res. accepted for publication
| + | 2013-07-05: |
| | | | |
| − | It also contains detailed informations about the program, examples and a discussion of limitations of the approach.
| + | * added two new labels: DOTA and K1 (pAcPhe based) |
| | | | |
| − | ===Contact===
| + | 2013-01-31: |
| − | hagelueken'at'pc.uni-bonn.de
| |
| | | | |
| − | ==Source Code==
| + | Version 1.1 in script repository. |
| − | <source lang="python">
| |
| − | """
| |
| − | ---mtsslWizard: spin_labeling plugin for PyMOL ---
| |
| − | Author : Gregor Hagelueken
| |
| − | Date : Jan 2012
| |
| − | Version : 1.0 | |
| − | Mail : gh50'at'st-andrews.ac.uk
| |
| | | | |
| − | mtsslWizard is a plugin for the PyMOL Molecular Graphics System.
| + | Some new features of v1.1: |
| − | The program is useful for in silico spin labeling of proteins with the spin label MTSSL in PyMOL. Also, distances between ensembles of two spin labels can be calculated and exported. | + | * Much faster. The algorithm was optimized and the speed of the program is now almost independent off the size of the molecule. Due to this, 'thorough search' is now set as default and the 'copy and move' mode has been dropped. |
| − | The program was tested with PyMOL versions 1.4.
| |
| | | | |
| − | Thanks to Jason Vertrees for help with the quick_dist function. Thanks to the author of the plane_wizard script for a nice example on how to program a PyMOL wizard.
| + | * New spin labels. The program now contains MTSSL, PROXYL and two labels for nucleic acids. Additional spin labels can be added upon request. |
| − |
| |
| − | Literature:
| |
| − | Hagelueken G, Ward R, Naismith JH, Schiemann O. MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. 2012. Appl. Mag. Res., accepted for publication.
| |
| − |
| |
| − | ----------------------------------------------------------------------
| |
| − | ----------------------------------------------------------------------
| |
| − |
| |
| − | """
| |
| − | import pymol
| |
| − | import numpy
| |
| − | import random, time, math
| |
| − | from pymol import cmd
| |
| − | from pymol.wizard import Wizard
| |
| − | from pymol import stored
| |
| − | from chempy import cpv
| |
| − | from pprint import pprint
| |
| − | from operator import itemgetter
| |
| | | | |
| | + | * Distances and histogram are directly copied to the clipboard or written out to a file as before. The result file contains 4 columns: 1-distances, 2-bins for histogram, 3-histogram frequencies, 4-histogram frequencies with highest value normalized to 1.0 for comparisons |
| | | | |
| − | default_thoroughness = "normal search"
| + | * Improved interface. Amongst other things, the clash control settings were simplified: There are now only two settings for vdW restraints: 'tight' and 'loose'. Also, only the average and c-Beta distances are displayed in the PyMOL viewer to avoid screen clutter (see screenshot). |
| − | default_cutoff = 3.4
| |
| − | default_clashes = 0
| |
| − | default_mode = 'Search'
| |
| − | default_rotamers = 'Unrestricted'
| |
| − | default_clashguard = 'on'
| |
| − | internalClash_cutoff = 2.5
| |
| | | | |
| − | def getAtypes(): #for internal clashGuard
| + | The new version was thoroughly tested. If you encounter any bugs or problems, please use the contact address below. |
| − | # 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
| |
| − | return [ 'N','C','O','CA','CB','SD','SG','CE','O1','N1','C2','C3','C4','C5','C6','C7','C8', 'C9', ]
| |
| | | | |
| − | def makeKnownExceptions(): #for clashGuard
| + | Version 1.0 is still available [https://github.com/Pymol-Scripts/Pymol-script-repo/blob/5935bdad82a480a4e004c8c902ef69dc75389ac8/plugins/mtsslWizard.py here]. |
| − | aTypes = getAtypes()
| |
| − | knownExceptions=[]
| |
| − | for x in range(len(aTypes)):
| |
| − | knownExceptions.append({})
| |
| − | for a in range(len(aTypes)):
| |
| − | for b in range(len(aTypes)):
| |
| − | knownExceptions[a][b] = 0
| |
| − | #pairs of atoms in this list are treated as exceptions for the internal clash guard
| |
| − | eList = [ [9,12], [0,1], [1,2], [1,3], [3,4], [4,6], [5,6], [5,7], [7,11], [8,9], [9,10], [9,13],
| |
| − | [10,11], [10,16], [10,17], [0,3], [0,4], [1,4], [2,3], [7,12], [8,10], [8,13],
| |
| − | [9,11], [9,14], [9,15], [9,16], [10,12], [10,13], [11,13], [11,12], [12,13], [12,14],
| |
| − | [13,14], [13,15], [12,15], [9,17], ]
| |
| − | for x in eList:
| |
| − | a,b = x
| |
| − | knownExceptions[a][b] = knownExceptions[b][a] = 1
| |
| | | | |
| − | return knownExceptions
| + | ==Program features== |
| | + | Spin labels can be attached to any position of a protein or nucleic acid just by pointing and clicking. The program then searches the conformational space of the label and determines which conformations of the label do not clash with the macromolecule. In "distance mode", distances between label ensembles can be determined and exported. In "distance map" mode, the program quickly estimates the inter-label distances between all possible spin pairs in a protein molecule. The result is saved as a color coded distance map. It is also possible to calculate difference distance maps between two conformations of a protein. |
| | | | |
| − | knownExceptions = makeKnownExceptions()
| + | Please reference: |
| | | | |
| − | def __init__(self):
| + | [http://www.springerlink.com/content/7148j5k2ppk475wu/ '''MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL''', Gregor Hagelueken, Richard Ward, James H. Naismith and Olav Schiemann, DOI: 10.1007/s00723-012-0314-0] |
| − | self.menuBar.addmenuitem('Wizard', 'command',
| |
| − | 'MtsslWizard',
| |
| − | label = 'MtsslWizard',
| |
| − | command = lambda s=self : open_wizard())
| |
| − | class MtsslWizard(Wizard):
| |
| − | def __init__(self):
| |
| − | Wizard.__init__(self)
| |
| − | print "MtsslWizard by gha. Please remove any solvent or unwanted heteroatoms before using the wizard!"
| |
| − |
| |
| − | #create contents of wizard menu
| |
| − | self.reset()
| |
| − | self.menu['mode'] = [
| |
| − | #[1, 'Stochastic','cmd.get_wizard().set_mode("Stochastic")'],
| |
| − | [1, 'Search','cmd.get_wizard().set_mode("Search")'],
| |
| − | [1, 'Measure','cmd.get_wizard().set_mode("Measure")'],
| |
| − | [1, 'Copy & Move','cmd.get_wizard().set_mode("Copy & Move")']
| |
| − | ]
| |
| − | #self.menu['rotamers'] =[
| |
| − | # [ 2, '\\955NOTE:\\559"Unrestricted" does not constrain the', ''],
| |
| − | # [ 2, '\\559chi angles at all. "Restricted" restrains the', ''],
| |
| − | # [ 2, '\\559chi angles to a rotamer library of MTSSL.', ''],
| |
| − | # [1, 'Unrestricted','cmd.get_wizard().set_rotamers("Unrestricted")'],
| |
| − | # [1, 'Restricted','cmd.get_wizard().set_rotamers("Restricted")']
| |
| − | # ]
| |
| − |
| |
| − | self.menu['thoroughness'] = [
| |
| − | [1, 'painstaking','cmd.get_wizard().set_thoroughness("painstaking")'],
| |
| − | [1, 'thorough search','cmd.get_wizard().set_thoroughness("thorough search")'],
| |
| − | [1, 'normal search','cmd.get_wizard().set_thoroughness("normal search")'],
| |
| − | [1, 'quick search','cmd.get_wizard().set_thoroughness("quick search")']
| |
| − | ]
| |
| − | self.menu['clashes'] = [
| |
| − | [ 2, '\\955NOTE:\\559This parameter should normally ', ''],
| |
| − | [ 2, '\\559be set to "0"! Use it with care and only ', ''],
| |
| − | [ 2, '\\559change it if conformational changes of ', ''],
| |
| − | [ 2, '\\559the protein are expected!!!', ''],
| |
| − | [1, '0 clashes','cmd.get_wizard().set_clashes(0)'],
| |
| − | [1, '1 clashes','cmd.get_wizard().set_clashes(1)'],
| |
| − | [1, '2 clashes','cmd.get_wizard().set_clashes(2)'],
| |
| − | [1, '3 clashes','cmd.get_wizard().set_clashes(3)'],
| |
| − | [1, '4 clashes','cmd.get_wizard().set_clashes(4)'],
| |
| − | [1, '5 clashes','cmd.get_wizard().set_clashes(5)']
| |
| − | ]
| |
| − |
| |
| − | self.menu['cutoff'] = [
| |
| − | [ 2, '\\955NOTE:\\559This parameter determines ', ''],
| |
| − | [ 2, '\\559the minimum allowed distance between', ''],
| |
| − | [ 2, '\\559label and protein atoms. Only change', ''],
| |
| − | [ 2, '\\559this parameter if conformational ch-', ''],
| |
| − | [ 2, '\\559anges at the labeling site are expected!', ''],
| |
| − | [1, '2.6 A','cmd.get_wizard().set_cutoff(2.6)'],
| |
| − | [1, '2.8 A','cmd.get_wizard().set_cutoff(2.8)'],
| |
| − | [1, '3.0 A','cmd.get_wizard().set_cutoff(3.0)'],
| |
| − | [1, '3.2 A','cmd.get_wizard().set_cutoff(3.2)'],
| |
| − | [1, '3.4 A','cmd.get_wizard().set_cutoff(3.4)'],
| |
| − | [1, '3.6 A','cmd.get_wizard().set_cutoff(3.6)'],
| |
| − | [1, '3.8 A','cmd.get_wizard().set_cutoff(3.8)']
| |
| − | ]
| |
| − |
| |
| − | self.menu['clashGuard'] = [
| |
| − | [ 2, '\\955NOTE:\\559The clashGuard checks for', ''],
| |
| − | [ 2, '\\559internal clashes of the label. ', ''],
| |
| − | [ 2, '\\559This should usually be set to "on"!', ''],
| |
| − | [1, 'on','cmd.get_wizard().set_clashGuard("on")'],
| |
| − | [1, 'off','cmd.get_wizard().set_clashGuard("off")']
| |
| − | ]
| |
| − |
| |
| − | self.menu['writeToFile'] = [
| |
| − | [1, 'no','cmd.get_wizard().set_writeToFile("no")'],
| |
| − | [1, 'yes','cmd.get_wizard().set_writeToFile("yes")']
| |
| − | ]
| |
| | | | |
| − | #some setter functions
| + | ==Screen shot== |
| − | def set_rotamers(self,rotamers):
| + | [[File:MtsslWizardv1-1.jpg]] |
| − | self.rotamers = rotamers
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def set_thoroughness(self,thoroughness):
| |
| − | self.thoroughness = thoroughness
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def set_writeToFile(self,writeToFile):
| |
| − | self.writeToFile = writeToFile
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def set_clashGuard(self,clashGuard):
| |
| − | self.clashGuard = clashGuard
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def set_cutoff(self,cutoff):
| |
| − | self.cutoff = cutoff
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def set_clashes(self,clashes):
| |
| − | self.clashes = clashes
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | #reset wizard to defaults
| |
| − | def reset(self):
| |
| − | self.toggleStatesCaption='Toggle states: ON'
| |
| − | self.start_time=time.time()
| |
| − | self.conformationList=[]
| |
| − | self.object_prefix = "mtsslWiz"
| |
| − | self.pick_count = 0
| |
| − | self.object_count = 0
| |
| − | self.allowedAngle=[False,False,False,False,False]
| |
| − | self.thoroughness = default_thoroughness
| |
| − | self.cutoff = default_cutoff
| |
| − | self.clashes = default_clashes
| |
| − | self.rotamers = default_rotamers
| |
| − | self.residue1_name = None
| |
| − | self.residue2_name = None
| |
| − | self.picked_object1 = None
| |
| − | self.picked_object2 = None
| |
| − | self.numberOfLabel = 0
| |
| − | self.mode=default_mode
| |
| − | self.writeToFile='no'
| |
| − | self.clashGuard=default_clashguard
| |
| − | self.selection_mode = cmd.get_setting_legacy("mouse_selection_mode")
| |
| − | cmd.set("mouse_selection_mode",1) # set selection mode to residue
| |
| − | cmd.deselect()
| |
| − | cmd.unpick()
| |
| − | cmd.delete(self.object_prefix + "*")
| |
| − | cmd.delete("sele*")
| |
| − | cmd.delete("_indicate*")
| |
| − | cmd.delete("pk*")
| |
| − | cmd.delete("*_tmp*")
| |
| − | cmd.refresh_wizard()
| |
| − |
| |
| − | def delete_all(self):
| |
| − | cmd.delete(self.object_prefix + "*")
| |
| − |
| |
| − | def set_mode(self, mode):
| |
| − | self.mode = mode
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def cleanup(self):
| |
| − | cmd.set("mouse_selection_mode",self.selection_mode) # restore selection mode
| |
| − | self.reset()
| |
| − | self.delete_all()
| |
| − |
| |
| − | def get_prompt(self):
| |
| − | self.prompt = None
| |
| − | if self.mode == 'Search' or 'Stochastic':
| |
| − | self.prompt = [ 'Select a residue to label...']
| |
| − | if self.pick_count == 0 and self.mode == 'Measure':
| |
| − | self.prompt = [ 'Select first label...']
| |
| − | if self.pick_count == 0 and self.mode == 'Copy & Move':
| |
| − | self.prompt = [ 'Select label to be copied & moved ...']
| |
| − | if self.pick_count == 1 and self.mode == 'Measure':
| |
| − | self.prompt = [ 'Select second label...']
| |
| − | if self.pick_count == 1 and self.mode == 'Copy & Move':
| |
| − | self.prompt = [ 'Select position for copied label ...']
| |
| − | return self.prompt
| |
| − |
| |
| − | def do_select(self, name):
| |
| − | # "edit" only this atom, and not others with the object prefix
| |
| − | try:
| |
| − | #cmd.edit("%s and not %s*" % (name, self.object_prefix))
| |
| − | self.do_pick(0)
| |
| − | except pymol.CmdException, pmce:
| |
| − | print pmce
| |
| − |
| |
| − |
| |
| − | def do_pick(self, picked_bond):
| |
| − | # this shouldn't actually happen if going through the "do_select"
| |
| − | if picked_bond:
| |
| − | self.error = "Error: please select bonds, not atoms"
| |
| − | print self.error
| |
| − | return
| |
| − | if self.pick_count == 0:
| |
| − | self.residue1_name = self.object_prefix + "_selectedResidue_" + str(self.pick_count)
| |
| − | # transfer the click selection to a named selection
| |
| − | cmd.select(self.residue1_name+"_tmp", "(sele)")
| |
| − | # delete the click selection
| |
| − | # highlight stuff
| |
| − | indicate_selection = "_indicate" + self.object_prefix + str(self.pick_count)
| |
| − | cmd.select(indicate_selection, self.residue1_name)
| |
| − | cmd.enable(indicate_selection)
| |
| − | # find the name of the object which contains the selection
| |
| − | new_name = None
| |
| − | obj_list = cmd.get_names('objects')
| |
| − | for object in obj_list:
| |
| − | if cmd.get_type(object)=="object:molecule":
| |
| − | if cmd.count_atoms("(%s and (sele))"%(object)):
| |
| − | self.picked_object1 = object
| |
| − | print self.picked_object1
| |
| − | break
| |
| − | src_frame = cmd.get_state()
| |
| − | if self.picked_object1 == None:
| |
| − | print " MtsslWizard: object not found."
| |
| − | self.pick_count += 1
| |
| − | if self.mode == 'Measure' or self.mode == 'Copy & Move':
| |
| − | #deselect before next pick
| |
| − | cmd.deselect()
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | elif self.pick_count == 1 and (self.mode == 'Measure' or self.mode == 'Copy & Move'):
| |
| − | self.residue2_name = self.object_prefix + "_selectedResidue_" + str(self.pick_count)
| |
| − | print self.residue2_name
| |
| − | # transfer the click selection to a named selection
| |
| − | cmd.select(self.residue2_name+"_tmp", "(sele)")
| |
| − | # highlight stuff
| |
| − | indicate_selection = "_indicate" + self.object_prefix + str(self.pick_count)
| |
| − | cmd.select(indicate_selection, self.residue2_name)
| |
| − | cmd.enable(indicate_selection)
| |
| − | # find the name of the object which contains the selection
| |
| − | new_name = None
| |
| − | obj_list = cmd.get_names('objects')
| |
| − | for object in obj_list:
| |
| − | if cmd.get_type(object)=="object:molecule":
| |
| − | if cmd.count_atoms("(%s and (sele))"%(object)):
| |
| − | self.picked_object2 = object
| |
| − | break
| |
| − | src_frame = cmd.get_state()
| |
| − | if self.picked_object2 == None:
| |
| − | print " MtsslWizard: object not found."
| |
| − | self.pick_count += 1
| |
| − | #deselect before next pick
| |
| − | self.run()
| |
| − | cmd.deselect()
| |
| − |
| |
| − | if self.mode == 'Search' or self.mode == 'Stochastic':
| |
| − | #print "increase numberOfLabel"
| |
| − | self.numberOfLabel += 1
| |
| − |
| |
| | | | |
| − | def run(self):
| + | ==Installation== |
| − | self.conformationList=[]
| + | Install the program by copying the code from the link above into an empty text file (e.g. "mtsslWizard.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. |
| − | my_view= cmd.get_view()
| + | Alternatively install the wizard via the plugins menu. |
| − | if self.pick_count == 1 and self.mode == 'Search':
| |
| − | print "\n\n\nNew run:\n"
| |
| − | mtsslify(self, self.picked_object1, self.residue1_name, self.numberOfLabel, self.thoroughness, self.cutoff, self.clashes)
| |
| − |
| |
| − | if self.pick_count == 1 and self.mode == 'Stochastic':
| |
| − | #testing selection...
| |
| − | print "\n\n\nNew run:\n"
| |
| − | stochasticMtsslify(self, self.picked_object1, self.residue1_name, self.numberOfLabel, self.thoroughness, self.cutoff, self.clashes)
| |
| − |
| |
| − | elif self.pick_count == 2 and self.mode == 'Measure':
| |
| − | print "\n\n\nNew run:\n"
| |
| − | quick_dist(self, self.picked_object1+" & name N1", self.picked_object2+" & name N1")
| |
| − |
| |
| − | elif self.pick_count == 2 and self.mode == 'Copy & Move':
| |
| − | print "\n\n\nNew run:\n"
| |
| − | copyAndMove(self.residue1_name, self.residue2_name, self.picked_object1, self.numberOfLabel)
| |
| − |
| |
| − | #some cleanup
| |
| − | self.pick_count = 0
| |
| − | cmd.delete("pk*")
| |
| − | cmd.delete("sele*")
| |
| − | cmd.delete("*_tmp*")
| |
| − | cmd.delete("_indicate*")
| |
| − | cmd.delete("labelEnvironment*")
| |
| − | self.cmd.refresh_wizard()
| |
| − | cmd.set_view(my_view)
| |
| − |
| |
| − |
| |
| − | def delete_last(self):
| |
| − | try:
| |
| − | print self.numberOfLabel
| |
| − | if self.numberOfLabel >= 1:
| |
| − | cmd.delete("mtsslWiz_"+str(self.numberOfLabel)+"*")
| |
| − | self.numberOfLabel-=1
| |
| − | except pymol.CmdException, pmce:
| |
| − | print pmce
| |
| − |
| |
| − | def toggle_states(self):
| |
| − | if cmd.get("all_states")=='on':
| |
| − | self.toggleStatesCaption='Toggle states: OFF'
| |
| − | cmd.set("all_states",0)
| |
| − | elif cmd.get("all_states")=='off':
| |
| − | self.toggleStatesCaption='Toggle states: ON'
| |
| − | cmd.set("all_states",1)
| |
| − | self.cmd.refresh_wizard()
| |
| − |
| |
| − | def get_panel(self):
| |
| − | if not ((self.mode == 'Measure') or (self.mode == 'Copy & Move')):
| |
| − | return [
| |
| − | [ 1, 'Mtssl Wizard',''],
| |
| − | [ 3, 'Mode: %s'%self.mode,'mode'],
| |
| − | #[ 3, 'Rotamers: %s'%self.rotamers,'rotamers'],
| |
| − | [ 3, 'Thoroughness: %s'%self.thoroughness,'thoroughness'],
| |
| − | [ 3, 'Cutoff: %3.1f A'%self.cutoff,'cutoff'],
| |
| − | [ 3, 'Clashes allowed: %i'%self.clashes,'clashes'],
| |
| − | #[ 3, 'Clash guard: %s'%self.clashGuard,'clashGuard'],
| |
| − | [ 3, 'Angles to file?: %s'%self.writeToFile,'writeToFile'],
| |
| − | [ 2, 'Go!','cmd.get_wizard().run()'],
| |
| − | [ 2, self.toggleStatesCaption,'cmd.get_wizard().toggle_states()'],
| |
| − | [ 2, 'Delete last label','cmd.get_wizard().delete_last()'],
| |
| − | [ 2, 'Reset','cmd.get_wizard().reset()'],
| |
| − | [ 2, 'Done','cmd.set_wizard()'],
| |
| − | ]
| |
| − | elif self.mode == 'Measure':
| |
| − | return [
| |
| − | [ 1, 'Mtssl Wizard',''],
| |
| − | [ 3, 'Mode: %s'%self.mode,'mode'],
| |
| − | [ 3, 'Distances to file?: %s'%self.writeToFile,'writeToFile'],
| |
| − | [ 2, 'Reset','cmd.get_wizard().reset()'],
| |
| − | [ 2, 'Done','cmd.set_wizard()']
| |
| − | ]
| |
| − | elif self.mode == 'Copy & Move':
| |
| − | return [
| |
| − | [ 1, 'Mtssl Wizard',''],
| |
| − | [ 3, 'Mode: %s'%self.mode,'mode'],
| |
| − | [ 2, 'Reset','cmd.get_wizard().reset()'],
| |
| − | [ 2, 'Done','cmd.set_wizard()']
| |
| − | ]
| |
| − |
| |
| | | | |
| − | def open_wizard():
| |
| − | wiz = MtsslWizard()
| |
| − | cmd.set_wizard(wiz)
| |
| | | | |
| − | cmd.extend('mtssl_wizard', open_wizard)
| + | mtsslWizard has been tested with PyMOL 1.7. It does not work with PyMOL 1.3. Here are manuals on how to install it on [[MAC_Install | Mac]] or [[Windows_Install | Windows]]. |
| | | | |
| − | def generateRandomChiAngle():
| + | Additional requirements: |
| − | chi=random.random()*360.0
| + | *SciPy has to be installed |
| − | return chi
| + | *Pyperclip or xerox have to be installed for the clipboard support. |
| | | | |
| | + | ====How people got it to run...==== |
| | + | *On a Mac it is easiest to install Pymol, Numpy, WxPython, Scipy and Matplotlib via fink. |
| | + | "I installed Pymol 1.5 via MacPorts. After putting pyperclip in the python 2.6 library folder, I then downloaded mtsslWizard and used the plugin installer to incorporate it. This was very simple and trouble-free." |
| | | | |
| − | def checkChi(self, chi, chiId):
| + | ==Usage== |
| − | #in free mode, all angles are allowed
| + | ===Labeling=== |
| − | if self.rotamers == 'Unrestricted':
| + | #Open a structure in PyMOL and remove any unwanted solvent molecules or ligands. |
| − | self.allowedAngle=[True, True, True, True, True]
| + | #Start the mtsslWizard via Wizards>mtsslWizard |
| − | #restrict chi angles to values from MMM rotamer library
| + | #If needed, vary the search parameters: "Speed" and "vdW restraints". 'Speed' determines how many conformations of the label will be generated and checked for clashes. The 'vdW restraints' parameter determines how stringent conformations are checked for clashes with the macromolecular surface. |
| − | elif self.rotamers == 'Restricted':
| + | #Click "Search conformers!" to start the calculation. |
| − | if chiId == 'chi1' and ((55.0 <= chi < 65.0) or (185.0 <= chi < 195.0) or (295.0 <= chi < 305.0)):
| |
| − | self.allowedAngle[0]=True
| |
| − | else:
| |
| − | self.allowedAngle[0]=False
| |
| − | if chiId == 'chi2' and ((60.0 <= chi < 90.0) or (165.0 <= chi < 195.0) or (272.0 <= chi < 303.0)):
| |
| − | self.allowedAngle[1]=True
| |
| − | else:
| |
| − | self.allowedAngle[1]=False
| |
| − | if chiId == 'chi3' and ((75.0 <= chi < 105.0) or (255.0 <= chi < 285.0)):
| |
| − | self.allowedAngle[2]=True
| |
| − | else:
| |
| − | self.allowedAngle[2]=False
| |
| − | if chiId == 'chi4' and ((70.0 <= chi < 90.0) or (165.0 <= chi < 195.0) or (272.0 <= chi < 292.0)):
| |
| − | self.allowedAngle[3]=True
| |
| − | else:
| |
| − | self.allowedAngle[3]=False
| |
| − | if chiId == 'chi5' and ((75.0 <= chi < 95.0) or (130.0 <= chi < 150.0) or (210.0 <= chi < 230.0) or (270.0 <= chi < 290.0)):
| |
| − | self.allowedAngle[4]=True
| |
| − | else:
| |
| − | self.allowedAngle[4]=False
| |
| | | | |
| | + | ===Distance calculation=== |
| | + | #change "Mode" to "Distance" |
| | + | #click the first and second ensemble |
| | + | #The program will now calculate all possible distances between the conformations in the two ensembles. If both ensembles have many conformations, this can take a couple of seconds! |
| | + | #The calculated distributions can be viewed with [[mtsslPlotter]]. |
| | | | |
| | + | Distances and a histogram are directly copied to the clipboard or written out to a file. The result file contains 4 columns: |
| | + | #distances |
| | + | #bins for histogram |
| | + | #histogram frequencies |
| | + | #histogram frequencies with highest value normalized to 1.0 for comparisons |
| | | | |
| − | def internalClash(self,selection): #this function checks for internal clashes of a new conformation
| |
| − | atoms=getAtypes()
| |
| − | for atom1 in range(len(atoms)-1):
| |
| − | for atom2 in range(atom1+1, len(atoms)):
| |
| − | # skip exceptions
| |
| − | if knownExceptions[atom1][atom2]==1:
| |
| − | #print "Excepted %s and %s" % (atoms[atom1], atoms[atom2])
| |
| − | continue
| |
| − | sel1, sel2 = selection + " & name " + atoms[atom1], selection + " & name " + atoms[atom2]
| |
| − | dist=cmd.dist("tmp_dist", sel1, sel2)
| |
| − | #cmd.select("isNeighbor", "(neighbor "+ sel1 + ") & (" + sel2 +")")
| |
| − | #count=cmd.count_atoms("isNeighbor")
| |
| − | #cmd.delete("isNeighbor")
| |
| − | cmd.delete("tmp_dist")
| |
| − |
| |
| − | if dist < internalClash_cutoff:
| |
| − | #print sel1, sel2, dist
| |
| − | return True
| |
| | | | |
| − | return False
| + | ===Distance maps=== |
| | + | #change "Mode" to "Distance map" |
| | + | #For a simple distance map: click twice on the protein of interest |
| | + | #For a difference distance map between two conformations of a protein: click once on each of the two conformations. For this feature to work properly, both conformations should have the same number of residues. This can be easily achieved using standard PyMOL commands. |
| | + | #The maps can be viewed with [[mtsslPlotter]] |
| | | | |
| − | def superpose(residue1,residue2):
| + | ==Reference== |
| − | #get the position of the selected residue's O atom
| + | If you find this program useful, please cite this paper: |
| − | stored.xyz = []
| |
| − | cmd.iterate_state(1,residue2+" & name O","stored.xyz.append([x,y,z])")
| |
| − | #print stored.xyz.pop(0)
| |
| − | if cmd.pair_fit(residue1+" & name CA",residue2+" & name CA",
| |
| − | residue1+" & name N", residue2+" & name N",
| |
| − | residue1+" & name C", residue2+" & name C",
| |
| − | residue1+" & name CB",residue2+" & name CB"):
| |
| − | #set the label's O atom to the stored position
| |
| − | cmd.alter_state(1,residue1+" & name O","(x,y,z)=stored.xyz.pop(0)")
| |
| − | return True
| |
| − | else:
| |
| − | return False
| |
| | | | |
| − | def copyAndMove(residue1_name, residue2_name, picked_object1, numberOfLabel):
| + | Hagelueken G, Ward R, Naismith JH, Schiemann O (2012) MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. Appl. Magn. Res. accepted for publication |
| − | label="mtssl_"+str(numberOfLabel)
| |
| − | cmd.copy(label+"_copied", picked_object1)
| |
| − | superpose(label+"_copied", residue2_name)
| |
| | | | |
| − | def mtsslify(self, selected_object, selected_residue, numberOfLabel, thoroughness, cutoff, maxClash): #generate and check conformations semi-systematically
| + | It also contains detailed informations about the program, examples and a discussion of limitations of the approach. |
| − | #max number of conformations
| |
| − | numberToFind=5000
| |
| − |
| |
| − | if thoroughness == 'painstaking':
| |
| − | repeat=500
| |
| − | elif thoroughness == 'thorough search':
| |
| − | repeat=100
| |
| − | elif thoroughness == 'normal search':
| |
| − | repeat=30
| |
| − | numberToFind=200
| |
| − | elif thoroughness == 'quick search':
| |
| − | repeat=10
| |
| − | numberToFind=100
| |
| − |
| |
| − | #generate the label and superpose onto selected position
| |
| − | generateMtssl(numberOfLabel)
| |
| − | label="mtssl_"+str(numberOfLabel)
| |
| − | print "Attempting superposition..."
| |
| − | if not superpose(label, selected_residue):
| |
| − | print "Superposition does not work. Glycine? Mutate to Ala first."
| |
| − | return
| |
| − | else:
| |
| − | print "Superposition worked!"
| |
| − |
| |
| − | #create object with only the atoms around the label to speed everything up
| |
| − | protein=selected_object+" &! "+ selected_residue + " within 15.0 of "+label
| |
| − | cmd.create("labelEnvironment_"+label,protein)
| |
| − |
| |
| − | #reset some counters and initialize variables
| |
| − | clashStateCounter=1
| |
| − | noClashStateCounter=1
| |
| − | internalClashCounter=1
| |
| − | snugglyFitStateCounter=1
| |
| − | chi1=0
| |
| − | chi2=0
| |
| − | chi3=0
| |
| − | chi4=0
| |
| − | chi5=0
| |
| − | if self.rotamers=='Unrestricted':
| |
| − | step=120
| |
| − | else:
| |
| − | step=30
| |
| − |
| |
| − | print "Trying to find conformations for label %s with thoroughness: %s" % (label, thoroughness)
| |
| − |
| |
| − | found=0
| |
| − | snugglyFitAtomCountList=[]
| |
| − | bestSnugglyFitAtomCount = 0
| |
| − | snugglyFitAtomCountSum = 0
| |
| − | bestSnugglyFitChiAngles = ""
| |
| − | cmd.select(label+"_r1a_head", label+" &! name N+CA+CB+O+C")
| |
| − |
| |
| − | #by running this repeatedly with different starting angles, the conformational space is sampled more quickly then by using a very small step size
| |
| − | x=0
| |
| − | for x in range (0,repeat):
| |
| − | self.allowedAngle=[False,False,False,False,False]
| |
| − | offset1=int(random.random()*step)
| |
| − | offset2=int(random.random()*step)
| |
| − | offset3=int(random.random()*step)
| |
| − | offset4=int(random.random()*step)
| |
| − | offset5=int(random.random()*step)
| |
| − | #chi1 loop
| |
| − | for chi1 in range(0+offset1,360+offset1,step):
| |
| − | checkChi(self, chi1, 'chi1')
| |
| − | if self.allowedAngle[0]==True and numberToFind > found:
| |
| − | cmd.set_dihedral(label+" & name N",label+" & name CA",label+" & name CB",label+" & name SG",chi1,state=1,quiet=1)
| |
| − | #check which atoms of protein lie within specified cutoff
| |
| − | cmd.create(label+"_clashAtoms", "labelEnvironment_"+label+" within "+str(cutoff)+" of "+label+" & name SG",1,clashStateCounter,1)
| |
| − | bumps=cmd.count_atoms(label+"_clashAtoms & state "+str(clashStateCounter))
| |
| − | cmd.delete(label+"_clashAtoms")
| |
| − | if bumps > 0:
| |
| − | print ".",
| |
| − | else:
| |
| − | #chi2 loop
| |
| − | for chi2 in range(0+offset2,360+offset2,step):
| |
| − | checkChi(self, chi2, 'chi2')
| |
| − | if self.allowedAngle[1]==True and numberToFind > found:
| |
| − | cmd.set_dihedral(label+" & name CA",label+" & name CB",label+" & name SG",label+" & name SD",chi2,state=1,quiet=1)
| |
| − | cmd.create(label+"_clashAtoms", "labelEnvironment_"+label+" within "+str(cutoff)+" of "+label+" & name SD",1,clashStateCounter,1)
| |
| − | bumps=cmd.count_atoms(label+"_clashAtoms & state "+str(clashStateCounter))
| |
| − | cmd.delete(label+"_clashAtoms")
| |
| − | if bumps>0:
| |
| − | print ".",
| |
| − | else:
| |
| − | #chi3 loop
| |
| − | for chi3 in range (0+offset3,360+offset3,step):
| |
| − | checkChi(self, chi3, 'chi3')
| |
| − | if self.allowedAngle[2]==True and numberToFind > found:
| |
| − | cmd.set_dihedral(label+" & name CB",label+" & name SG",label+" & name SD",label+" & name CE",chi3,state=1,quiet=1)
| |
| − | cmd.create(label+"_clashAtoms", "labelEnvironment_"+label+" within "+str(cutoff)+" of "+label+" & name CE",1,clashStateCounter,1)
| |
| − | bumps=cmd.count_atoms(label+"_clashAtoms & state "+str(clashStateCounter))
| |
| − | cmd.delete(label+"_clashAtoms")
| |
| − | if bumps>0:
| |
| − | print ".",
| |
| − | else:
| |
| − | #chi4 loop
| |
| − | for chi4 in range (0+offset4,360+offset4,step):
| |
| − | checkChi(self, chi4, 'chi4')
| |
| − | if self.allowedAngle[3]==True and numberToFind > found:
| |
| − | cmd.set_dihedral(label+" & name SG",label+" & name SD",label+" & name CE",label+" & name C3",chi4,state=1,quiet=1)
| |
| − | #cmd.select(label+"_r1a_head", label+" &! name N+CA+CB+O+C")
| |
| − | cmd.create(label+"_clashAtoms", "labelEnvironment_"+label+" within "+str(cutoff)+" of "+label+"_r1a_head",1,clashStateCounter,1)
| |
| − | bumps=cmd.count_atoms(label+"_clashAtoms & state "+str(clashStateCounter))
| |
| − | cmd.delete(label+"_clashAtoms")
| |
| − | if bumps>0:
| |
| − | print ".",
| |
| − | else:
| |
| − | #chi5 loop
| |
| − | for chi5 in range (0+offset5,360+offset5,step):
| |
| − | checkChi(self, chi5, 'chi5')
| |
| − | if self.allowedAngle[4]==True and numberToFind > found:
| |
| − | cmd.set_dihedral(label+" & name SD",label+" & name CE",label+" & name C3",label+" & name C4",chi5,state=1,quiet=1)
| |
| − | cmd.create(label+"_clashAtoms", "labelEnvironment_"+label+" within "+str(cutoff)+" of "+label+"_r1a_head",1,clashStateCounter,1)
| |
| − | bumps=cmd.count_atoms(label+"_clashAtoms & state "+str(clashStateCounter))
| |
| − | cmd.delete(label+"_clashAtoms")
| |
| − | if bumps<=maxClash:
| |
| − | found+=1
| |
| − | if self.clashGuard=="on" and internalClash(self, label):
| |
| − | print found,
| |
| − | cmd.create(label+"_internal_clash", label, 1, internalClashCounter)
| |
| − | internalClashCounter+=1
| |
| − | else:
| |
| − | cmd.create(label+"_no_clash", label, 1, noClashStateCounter)
| |
| − | cmd.select("snugglyFitTest", "labelEnvironment_"+label+" & ("+label+"_r1a_head around 4.5)")
| |
| − | snugglyFitAtomCount=cmd.count_atoms("snugglyFitTest")
| |
| − | cmd.delete("snugglyFitTest")
| |
| − | #make entry in conformationList
| |
| − | thisConformation=[chi1,chi2,chi3,chi4,chi5,snugglyFitAtomCount,noClashStateCounter]
| |
| − | self.conformationList.append(thisConformation)
| |
| − | print found,
| |
| − | noClashStateCounter+=1
| |
| − | else:
| |
| − | print ".",
| |
| − | x+=1
| |
| − | #carriage return
| |
| − | print "\n"
| |
| − | if len(self.conformationList) > 0:
| |
| − | if self.thoroughness == 'painstaking':
| |
| − | createSnugglyFitConformations(label,self.conformationList)
| |
| − | polarPlot(self, label, self.conformationList)
| |
| − | finalCosmetics(self, label,numberOfLabel,found,thoroughness, maxClash,cutoff)
| |
| − | print "Done!"
| |
| − | | |
| − | | |
| − | def createSnugglyFitConformations(label,conformationList): #select conformations with the best fit to the molecular surface, rank them and create an object
| |
| − | print "Snuggliest fit(s):"
| |
| − | #calculate average atom count of all conformations
| |
| − | atomCountSum=0
| |
| − | snugglyFitList=[]
| |
| − | bestSnugglyFitAtomCount=0
| |
| − | for x in range (0,len(conformationList)):
| |
| − | thisConformation=conformationList[x]
| |
| − | atomCountSum+=thisConformation[5]
| |
| − | if thisConformation[5] > bestSnugglyFitAtomCount:
| |
| − | bestSnugglyFitAtomCount=thisConformation[5]
| |
| − | averageAtomCount=atomCountSum/len(conformationList)
| |
| − | #generate snugglyFitList: take only those conformations whose atom count is > 0.75 of top peak and higher than the average
| |
| − | counter=1
| |
| − | for x in range (0,len(conformationList)):
| |
| − | thisConformation=conformationList[x]
| |
| − | if thisConformation[5] > 0.75 * bestSnugglyFitAtomCount and thisConformation[5] > averageAtomCount:
| |
| − | snugglyFitList.append({'chi1':thisConformation[0],
| |
| − | 'chi2':thisConformation[1],
| |
| − | 'chi3':thisConformation[2],
| |
| − | 'chi4':thisConformation[3],
| |
| − | 'chi5':thisConformation[4],
| |
| − | 'vdw':thisConformation[5],
| |
| − | 'state':thisConformation[6]})
| |
| − | cmd.create(label+"_snuggly", label+"_no_clash", thisConformation[6], counter)
| |
| − | counter+=1
| |
| − | #sort snugglyFitList so that best fitting conformation is on top
| |
| − | snugglyFitList = sorted(snugglyFitList, key=itemgetter('vdw'))
| |
| − | snugglyFitList.reverse()
| |
| − | #print out the list
| |
| − | if len(snugglyFitList)>0:
| |
| − | for i in range (0,len(snugglyFitList)):
| |
| − | print "Conformation %i: \t\t%i \t\t\t vdW contacts" %(snugglyFitList[i]['state'],snugglyFitList[i]['vdw'])
| |
| − | print "Average vdW contacts of all possible conformations: ",averageAtomCount
| |
| − |
| |
| − |
| |
| − |
| |
| − | | |
| − | def finalCosmetics(self, label, numberOfLabel, found, thoroughness,maxClash,cutoff): #make everything look nice
| |
| − | if found >= 1:
| |
| − | print "Found %i conformations." %found
| |
| − | #show all conformations and do some coloring
| |
| − | cmd.set("all_states",1)
| |
| − | self.toggleStatesCaption='Toggle states: ON'
| |
| − | cmd.color("blue",label+"_no_clash")
| |
| − | cmd.color("red", "name O1")
| |
| − | cmd.disable(label)
| |
| − | cmd.disable(label+"_internal_clash")
| |
| − | cmd.show("spheres", "name O1")
| |
| − | cmd.set("sphere_scale", "0.2")
| |
| − | identifierLabel="%s|%s|%s|%1.2f|%i|%s" %(label,self.mode, self.rotamers, cutoff, maxClash, thoroughness)
| |
| − | print identifierLabel
| |
| − | cmd.pseudoatom(label+"_label", label)
| |
| − | cmd.label(label+"_label", `identifierLabel`)
| |
| − | cmd.show("label")
| |
| − | cmd.group("mtsslWiz_"+str(numberOfLabel), label+"*"+","+"labelEnvironment_"+label)
| |
| − | #cmd.hide("label")
| |
| − | else:
| |
| − | print "Sorry, I did not find anything. Your options are:\n1) Try again or\n2) Try to increase 'thoroughness' (now: "+str(thoroughness)+"), \nincrease'maxClash' (now: "+str(maxClash)+") or \ndecrease cutoff (now: "+str(cutoff)+")."
| |
| − | cmd.delete(label+"*")
| |
| − | print label
| |
| − |
| |
| − | | |
| − | def polarPlot(self,label,conformationList): #generate file for polar plot. Use polar_plot.py to plot it.
| |
| − | if self.writeToFile=='yes':
| |
| − | chiFile=open(label+"_chiFile.txt", 'w')
| |
| − | for x in range (0, len(conformationList)):
| |
| − | thisConformation=conformationList[x]
| |
| − | chiFile.write("%s %s %s %s %s\n" % (thisConformation[0],thisConformation[1],thisConformation[2],thisConformation[3],thisConformation[4]))
| |
| − | chiFile.close()
| |
| − |
| |
| | | | |
| | + | ==Older versions== |
| | + | *[https://github.com/Pymol-Scripts/Pymol-script-repo/blob/5935bdad82a480a4e004c8c902ef69dc75389ac8/plugins/mtsslWizard.py Version 1.0] |
| | | | |
| − | #calculate distances between all possible conformations
| + | ==Contact== |
| − | def quick_dist(self, s1, s2, inFile=None):
| + | hagelueken'at' uni-bonn.de |
| − | if self.writeToFile=='yes':
| |
| − | filename="distances_%s-%s.txt" %(self.picked_object1,self.picked_object2)
| |
| − | filename.replace(' ', '_')
| |
| − | f = open(filename, 'w')
| |
| − | s=""
| |
| − | s+="DIST\n"
| |
| − | distanceList=[]
| |
| − | #find the amount of conformations
| |
| − | m1States=cmd.count_states(s1)
| |
| − | m2States=cmd.count_states(s2)
| |
| − | #do the distance calculation for all states
| |
| − | for i in range (1,m1States+1):
| |
| − | print ".",
| |
| − | for j in range (1,m2States+1):
| |
| − | m1 = cmd.get_model(s1,i)
| |
| − | m2 = cmd.get_model(s2,j)
| |
| − | for c1 in range(len(m1.atom)):
| |
| − | for c2 in range(len(m2.atom)):
| |
| − | #current distance
| |
| − | currentDistance=math.sqrt(sum(map(lambda f: (f[0]-f[1])**2, zip(m1.atom[c1].coord,m2.atom[c2].coord))))
| |
| − | distanceList.append(currentDistance)
| |
| − | #print out table
| |
| − | s+="%3.2f\n" % (currentDistance)
| |
| − |
| |
| − | cmd.distance(s1,s2)
| |
| − | #calculate cbeta for comparison
| |
| − | s1=self.picked_object1+" & name CB"
| |
| − | s2=self.picked_object2+" & name CB"
| |
| − | m1 = cmd.get_model(s1,1)
| |
| − | m2 = cmd.get_model(s2,1)
| |
| − | for c1 in range(len(m1.atom)):
| |
| − | for c2 in range(len(m2.atom)):
| |
| − | cBeta=math.sqrt(sum(map(lambda f: (f[0]-f[1])**2, zip(m1.atom[c1].coord,m2.atom[c2].coord))))
| |
| − | s+="\n"
| |
| − | cmd.dist("cB_"+self.picked_object1+"_"+self.picked_object2, self.picked_object1+" & name CB", self.picked_object2+" & name CB")
| |
| − |
| |
| − | if self.writeToFile=='yes':
| |
| − | f.write(s)
| |
| − | f.close()
| |
| − | print calculateStatistics(distanceList)
| |
| − | print "Cbeta distance: %3.1f" %cBeta
| |
| − |
| |
| − |
| |
| − | def calculateStatistics(distanceList_strings): #create distance histogram...
| |
| − | statisticsResult=""
| |
| − | #easy statistics
| |
| − | distanceList=sorted([float(x) for x in distanceList_strings])
| |
| − | average = sum(distanceList)/len(distanceList)
| |
| − | median = distanceList[len(distanceList)/2]
| |
| − | longest = distanceList[len(distanceList)-1]
| |
| − | shortest = distanceList[0]
| |
| − | #generate histogram
| |
| − | bins=[]
| |
| − | bins.append(shortest)
| |
| − | binSize=(longest-shortest)/10
| |
| − | for i in range (1, 10, 1):
| |
| − | bins.append(shortest + i * binSize)
| |
| − | hist,bin_edges=numpy.histogram(distanceList,bins=bins,normed=True)
| |
| − | histDict={}
| |
| − | histZoomFactor=100
| |
| − | max_freq=0
| |
| − | #convert histogram to dictionary for easier generation of text histogram
| |
| − | print
| |
| − | for i in range(0,len(hist),1):
| |
| − | histDict[i]=int(round(hist[i]*histZoomFactor))
| |
| − | #print histDict[i]
| |
| − | if histDict[i] > max_freq:
| |
| − | max_freq = histDict[i]
| |
| − | #print out stuff
| |
| − | statisticsResult+="Distance histogram:\n"
| |
| − | for i in range(max_freq, -1, -1):
| |
| − | for bin in sorted(histDict.keys()):
| |
| − | if histDict[bin] >= i:
| |
| − | statisticsResult+='#'
| |
| − | else:
| |
| − | statisticsResult+=" "
| |
| − | statisticsResult+="\n"
| |
| − | statisticsResult+= "Average distance: %3.2f\n" %average
| |
| − | statisticsResult+= "Median of distribution: %3.2f\n" %median
| |
| − | statisticsResult+= "Shortest distance: %3.2f\n" % shortest
| |
| − | statisticsResult+= "Longest distance: %3.2f" %longest
| |
| − | return statisticsResult
| |
| | | | |
| − | def generateMtssl(numberOfLabel): #create a mtssl label object.
| + | ==Acknowledgement== |
| − | cmd.read_pdbstr("""HEADER MTSSL\n
| + | Thanks to Jason Vertrees and Thomas Holder for some programming tips. |
| − | COMPND coordinates of R1A from program: libcheck\n
| |
| − | CRYST1 100.000 100.000 100.000 90.00 90.00 90.00 P 1\n
| |
| − | SCALE1 0.010000 0.000000 0.000000 0.00000\n
| |
| − | SCALE2 0.000000 0.010000 0.000000 0.00000\n
| |
| − | SCALE3 0.000000 0.000000 0.010000 0.00000\n
| |
| − | ATOM 1 N R1A A 1 0.201 -0.038 -0.149 1.00 20.00 N\n
| |
| − | ATOM 2 CA R1A A 1 1.258 1.007 -0.271 1.00 20.00 C\n
| |
| − | ATOM 4 CB R1A A 1 2.056 0.796 -1.554 1.00 20.00 C\n
| |
| − | ATOM 7 SG R1A A 1 3.667 1.492 -1.387 1.00 20.00 S\n
| |
| − | ATOM 8 SD R1A A 1 4.546 1.587 -3.180 1.00 20.00 S\n
| |
| − | ATOM 9 CE R1A A 1 5.573 3.020 -3.244 1.00 20.00 C\n
| |
| − | ATOM 12 C3 R1A A 1 6.644 3.007 -4.321 1.00 20.00 C\n
| |
| − | ATOM 13 C4 R1A A 1 7.349 4.109 -4.593 1.00 20.00 C\n
| |
| − | ATOM 15 C5 R1A A 1 8.361 3.885 -5.680 1.00 20.00 C\n
| |
| − | ATOM 16 C7 R1A A 1 9.758 4.118 -5.119 1.00 20.00 C\n
| |
| − | ATOM 20 C6 R1A A 1 8.092 4.808 -6.859 1.00 20.00 C\n
| |
| − | ATOM 24 N1 R1A A 1 8.144 2.482 -5.989 1.00 20.00 N\n
| |
| − | ATOM 25 O1 R1A A 1 8.792 1.889 -6.835 1.00 20.00 O\n
| |
| − | ATOM 26 C2 R1A A 1 7.092 1.857 -5.197 1.00 20.00 C\n
| |
| − | ATOM 27 C8 R1A A 1 7.642 0.712 -4.360 1.00 20.00 C\n
| |
| − | ATOM 31 C9 R1A A 1 5.961 1.403 -6.123 1.00 20.00 C\n
| |
| − | ATOM 35 C R1A A 1 0.670 2.388 -0.261 1.00 20.00 C\n
| |
| − | ATOM 36 O R1A A 1 -0.298 2.670 -0.967 1.00 20.00 O\n""","mtssl_"+ str(numberOfLabel))
| |
| − | </source>
| |