This is a read-only mirror of pymolwiki.org
MtsslTrilaterate
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/mtsslTrilaterate.py |
| Author(s) | Dinar Abdullin |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
mtsslTrilaterate
mtsslTrilaterate is a PyMOL plugin for the trilateration of objects based on distance restraints.
The mtsslTrilaterate program calculates the coordinates of an object which is defined by the distances from this object to several known objects in a 3D space. This program can be used to localize spin centers in biomolecules by measuring the distances between these metal ions and artificially attached spin labels. The mtsslTrilaterate program is a part of the mtsslSuite package and it is optimized to work in pair with mtsslWizard program. The mtsslTrilaterate program is a python code and it works as a plugin for the PyMOL (www.pymol.org) program. It was tested on the Windows 7 64 bit platform. Due to some problems with the wx library in PyMOL the program temporarily does not work on the Mac platform.
News
2013-07-09:
Version 1.0 in repository.
If you use this program, please reference:
Screen shot
Installation
1. mtsslTrilaterate requires python 2.7 or higher to be installed. It can be downloaded free of charge from official web site http://www.python.org/. The program also uses wx and numpy modules. If the basic version of Python does not contain these modules they should be installed subsequently (e.g. from http://www.lfd.uci.edu/~gohlke/pythonlibs/). 2. mtsslTrilaterate requires PyMOL version compatible with Python 2.7. 3. Save trilater.py file somewhere in your computer. 4. Run PyMOL. In the Plugin menu select Plugin Manager. Then open the Install New Plugin page and click on Choose file. Select the trilater.py file and choose a plugin directory. If installation is finished successfully a message “Plugin Trilateration has been installed” would appear. 5. Run mtsslTrilaterate from the Plugin menu.
mtsslTrilaterate has been tested with PyMOL 1.5.
Additional requirements:
- wx
- NumPy
Trilateration
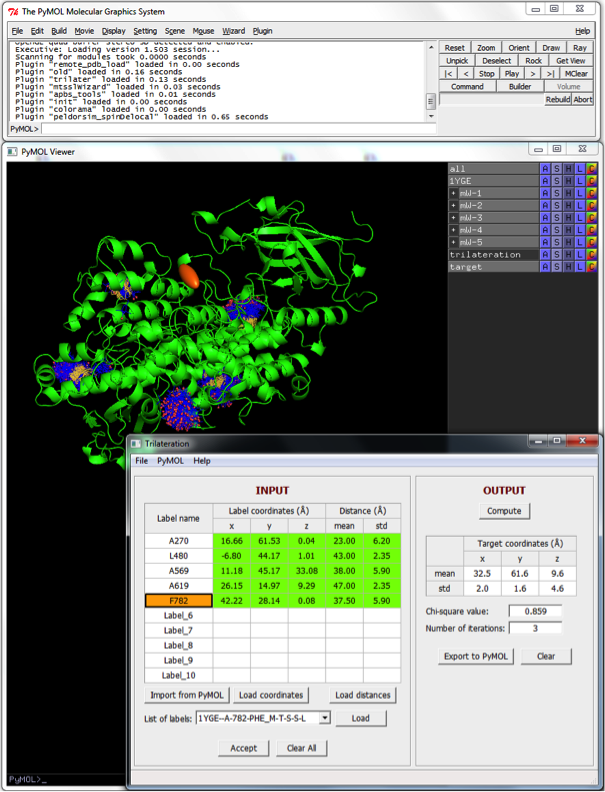
User interface
The graphical user interface of mtsslTrilaterate is presented on Figure 1. It contains a small menu and a main working window. The main window consists of 2 main panes called “INPUT” and “OUTPUT”. The “INPUT” pane represents all input data as a table. Each row of this table is related to a certain label (spin label) that was introduced to the structural model. The identifiers of these labels are listed in the first column. The following three columns correspond to the average coordinate (x, y, z) of the label. The last two columns represent the mean value and the standard deviation of the distances between the particular label and a target.
1. Load biomolecule structure to current PyMOL session. It can be done either in PyMOL selecting File → Open or in mtsslTrilaterate selecting PyMOL → Load structure to PyMOL.
2. Create spin labels by means of mtsslWizard selecting in PyMOL Wizard → mtsslWizard. This wizard performs site-directed spin labelling in silico (look http://pymolwiki.org/index.php/MtsslWizard for more information).
3. The coordinates of the selected label can be entered in three different ways. Firstly, they can be entered manually to the table. Secondly, the parameters related to a particular spin label can be imported from the current PyMOL session by pushing the “Import from PyMOL” button. The identifiers and average coordinates of spin labels that were generated inside PyMOL beforehand are in this way imported into the program and can be assigned to rows in the data table. Thirdly, the label coordinates can be loaded from an ASCII data file by selecting the label in the table and then clicking the “Load coordinates” button. The data file should contain 3 columns corresponding to x, y, z coordinates. If the label is flexible (i.e. has a set of the coordinates) an average label coordinates will be calculated after loading the data file.
4. The distance between the selected label and the target can be entered manually to the corresponding fields of the table or loaded from an ASCII data file clicking the “Load distances” button. This way an interspin distance distribution generated within the DeerAnalysis program (Jeschke, G.; Chechik, V.; Ionita, P.; Godt, A.; Zimmermann, H.; Banham, J.; Timmel, C. R.; Hilger, D.; Jung, H. Appl. Magn. Reson. 2006, 30, 473) can be imported to the program.
5. If all input data entered press “Accept” button. At this stage the program checks if at least four spin labels were described completely. Four spin labels is a minimal number of labels that allows to solve the trilateration problem.
6. Click “Compute” button. After calculations, the coordinates of the target and their standard deviations should appear in the “OUTPUT” pane. The program also displays an obtained χ2 value and a number of iterations which was required to reach this χ2 value.
7. Push Export to PyMOL button to get a graphical representation of trilateration in the current PyMOL session. Two PyMOL objects, called “trilateration” and “target”, will be created inside PyMOL (Figure 1). The “trilateration” object represents a ‘trilateration-sphere’ plotted around each used spin label. The radius of each sphere equals to the average distance between the corresponding spin label and the unlocalized spin center. The space where these spheres intersect each other determines the location of the spin center. This space is depicted by the “target” ellipsoid. The center of the ellipsoid corresponds to the most probable coordinates of the spin center, whereas the legths of its semi-principal axes equal to the obtained standard errors of these coordinates. The scale of semi-principal axes, i.e. the confidence level of the solution, can be incresed in the File → Preferences → Output pane.
8. Click “Save as” button or select File → Save as in the File menu to save current section as a ASCII data file. Afterwards this session can be reinitialized by opening this data file through File → Open.
The “OUTPUT” pane is used to displays the results of the trilateration process. After the calculation is started by pushing the “Compute” button, the program solves the trilateration problem and displays the most probable coordinates of the target and their standard errors.
The File menu contains the Open, Save as, and Close commands to work with data files. It also incorporates the Preferences window which allows user to format the input and output data of the program as well as to adjuxt some calculation parameters.
The PyMOL menu allows user to open and close the PyMOL data files within the program.
Reference
If you find this program useful, please cite this paper:
It also contains detailed informations about the program, examples and a discussion of limitations of the approach.
Contact
abdullin'at'pc.uni-bonn.de
Acknowledgement
Thanks to Jason Vertrees and Thomas Holder for some programming tips.