This is a read-only mirror of pymolwiki.org
Distance
DESCRIPTION
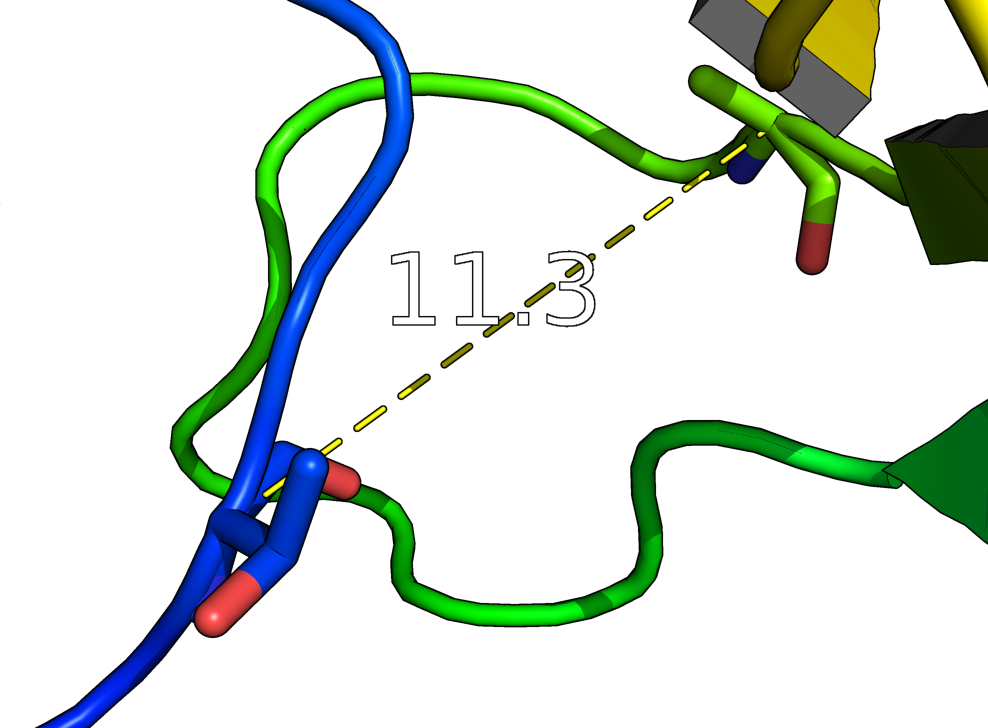
distance creates a new distance object between two selections. It will display all distances within the cutoff.
USAGE
distance (selection1), (selection2)
# example
dist i. 158 and n. CA, i. 160 and n. CA
distance name = (selection1), (selection2) [,cutoff [,mode] ]
- name = name of distance object
- selection1, selection2 = atom selections
- cutoff = maximum distance to display
- mode = 0 (default)
PYMOL API
cmd.distance( string name, string selection1, string selection2,
string cutoff, string mode )
# returns the average distance between all atoms/frames
NOTES
The distance wizard makes measuring distances easier than using the "dist" command for real-time operations.
"dist" alone will show distances between selections (pk1) and (pk1), which can be set using the PkAt mouse action (usually CTRL-middle-click).
EXAMPLES
- Get and show the distance from residue 10's alpha carbon to residue 40's alpha carbon in 1ESR:
# make the first residue 0.
zero_residues 1esr, 0
distance i. 10 and n . CA, i. 40 and n. CA
- Get and show the distance from residue 10's alpha carbon to residue 35-42's alpha carbon in 1ESR:
# make the first residue 0.
zero_residues 1esr, 0
distance i. 10 and n . CA, i. 35-42 and n. CA
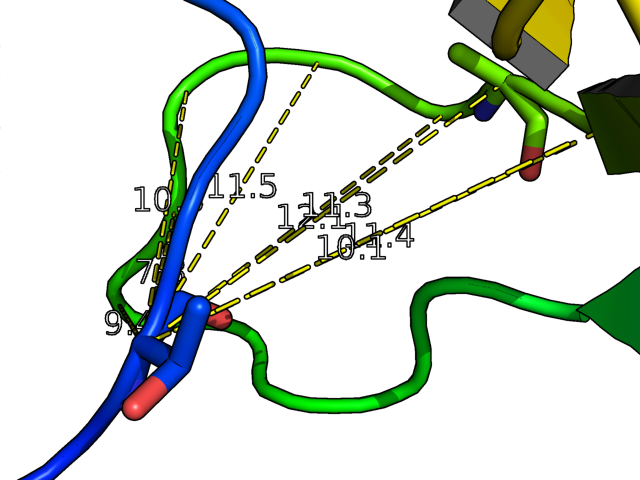
- This neat example shows how to create distance measurements from an atom in a molecule to all other atoms in the molecule (since PyMol supports wildcards).
cmd.dist("(/mol1///1/C)","(/mol1///2/C*)")
or written without the PyMolScript code,
dist /mol1///1/C, /mol1///2/C*
- Create multiple distance objects
for at1 in cmd.index("resi 10"): \
for at2 in cmd.index("resi 11"): \
cmd.dist(None, "%s`%d"%at1, "%s`%d"%at2)