This is a read-only mirror of pymolwiki.org
Difference between revisions of "Dehydron"
| Line 2: | Line 2: | ||
|type = plugin | |type = plugin | ||
|filename = plugins/dehydron.py | |filename = plugins/dehydron.py | ||
| − | |author = Osvaldo Martin | + | |author = [[User:aloctavodia|Osvaldo Martin]] |
|license = GPL | |license = GPL | ||
}} | }} | ||
== Introduction == | == Introduction == | ||
| − | A dehydron calculator plugin for PyMOL <br> | + | A dehydron calculator plugin for PyMOL. This plugin calculates dehydrons and display them onto the protein structure. |
| − | + | ||
| + | A dehydron is a main chain hydrogen bond incompletely shielded from water attack. For a brief introduction to the dehydron concept, please read http://en.wikipedia.org/wiki/Dehydron | ||
| + | |||
| + | == Installation == | ||
| + | === Linux === | ||
| + | This plugin is ready "out-of-box" for Linux users through the project [[git_intro | Pymol-script-repo]] | ||
| + | |||
| + | === Windows === | ||
| + | This plugin is ready "out-of-box" for win users through the project [[git_intro | Pymol-script-repo]] | ||
| + | |||
| + | == Usage == | ||
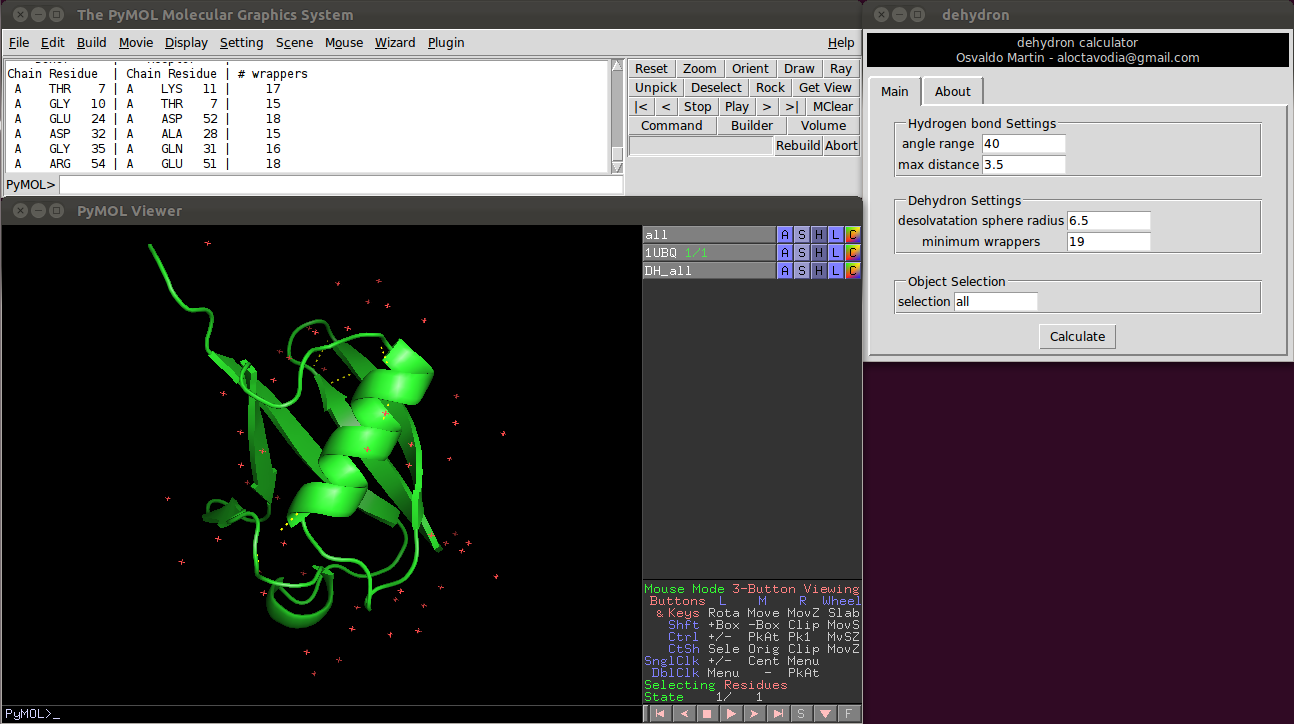
| + | [[Image:Dehydrons.png|400px|thumb|Dehydrons calculated and displayed in PyMOL.]] | ||
| + | There are four parameters the user can change:<br> | ||
| + | <br> | ||
| + | Two of them control the hydrogen bonds detection.<br> | ||
| + | * Angle range: deviation in degrees from the optimal hydrogen bond angle (C=0 N-H).<br> | ||
| + | * Max distance: maximum donor-aceptor distance.<br> | ||
| + | <br> | ||
| + | And the other two control the dehydron detection<br> | ||
| + | * Desolvatation sphere radius: this parameter controls the radius of the two spheres centered at the ca carbon of the donor and acceptor residues. A dehydron is defined by the number of "wrappers" inside this two spheres.<br> | ||
| + | * Cut-off wrappers: a hydrogen bond surrounded with less "wrappers" than "Cut-off wrappers" is a dehydron.<br> | ||
| + | |||
| + | A wrapper is defined as a carbon atom not bonded directly to an oxygen or nitrogen atom, i.e. a non-polar carbon atom. | ||
| + | |||
| + | == Acknowledgement == | ||
| + | The H-bond detection code is based on the list_mc_hbonds.py script from Robert L. Campbell http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/ | ||
| + | |||
| + | == References == | ||
| + | Citation for Dehydrons:<br> | ||
| + | Fernández A. and Scott R.; "Adherence of packing defects in soluble proteins", Phys. Rev. Lett. 91, 018102 (2003). | ||
| + | |||
| + | Fernández A., Rogale K., Scott R. and Scheraga H.A.; "Inhibitor design by wrapping packing defects in HIV-1 proteins", PNAS, 101, 11640-45 (2004). | ||
| + | |||
| + | Fernández A. "Transformative Concepts for Drug Design: Target Wrapping" (ISBN 978-3642117916), Springer-Verlag, Berlin, Heidelberg (2010). | ||
| − | |||
| − | |||
[[Category:Plugins]] | [[Category:Plugins]] | ||
[[Category:Pymol-script-repo]] | [[Category:Pymol-script-repo]] | ||
Revision as of 14:46, 18 January 2012
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/dehydron.py |
| Author(s) | Osvaldo Martin |
| License | GPL |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
A dehydron calculator plugin for PyMOL. This plugin calculates dehydrons and display them onto the protein structure.
A dehydron is a main chain hydrogen bond incompletely shielded from water attack. For a brief introduction to the dehydron concept, please read http://en.wikipedia.org/wiki/Dehydron
Installation
Linux
This plugin is ready "out-of-box" for Linux users through the project Pymol-script-repo
Windows
This plugin is ready "out-of-box" for win users through the project Pymol-script-repo
Usage
There are four parameters the user can change:
Two of them control the hydrogen bonds detection.
- Angle range: deviation in degrees from the optimal hydrogen bond angle (C=0 N-H).
- Max distance: maximum donor-aceptor distance.
And the other two control the dehydron detection
- Desolvatation sphere radius: this parameter controls the radius of the two spheres centered at the ca carbon of the donor and acceptor residues. A dehydron is defined by the number of "wrappers" inside this two spheres.
- Cut-off wrappers: a hydrogen bond surrounded with less "wrappers" than "Cut-off wrappers" is a dehydron.
A wrapper is defined as a carbon atom not bonded directly to an oxygen or nitrogen atom, i.e. a non-polar carbon atom.
Acknowledgement
The H-bond detection code is based on the list_mc_hbonds.py script from Robert L. Campbell http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/
References
Citation for Dehydrons:
Fernández A. and Scott R.; "Adherence of packing defects in soluble proteins", Phys. Rev. Lett. 91, 018102 (2003).
Fernández A., Rogale K., Scott R. and Scheraga H.A.; "Inhibitor design by wrapping packing defects in HIV-1 proteins", PNAS, 101, 11640-45 (2004).
Fernández A. "Transformative Concepts for Drug Design: Target Wrapping" (ISBN 978-3642117916), Springer-Verlag, Berlin, Heidelberg (2010).