This is a read-only mirror of pymolwiki.org
Contact map visualizer
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/contact_map_visualizer.py |
| Author(s) | Venkatramanan Krishnamani |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
Enhanced version of this plugin is now available at CMPyMOL
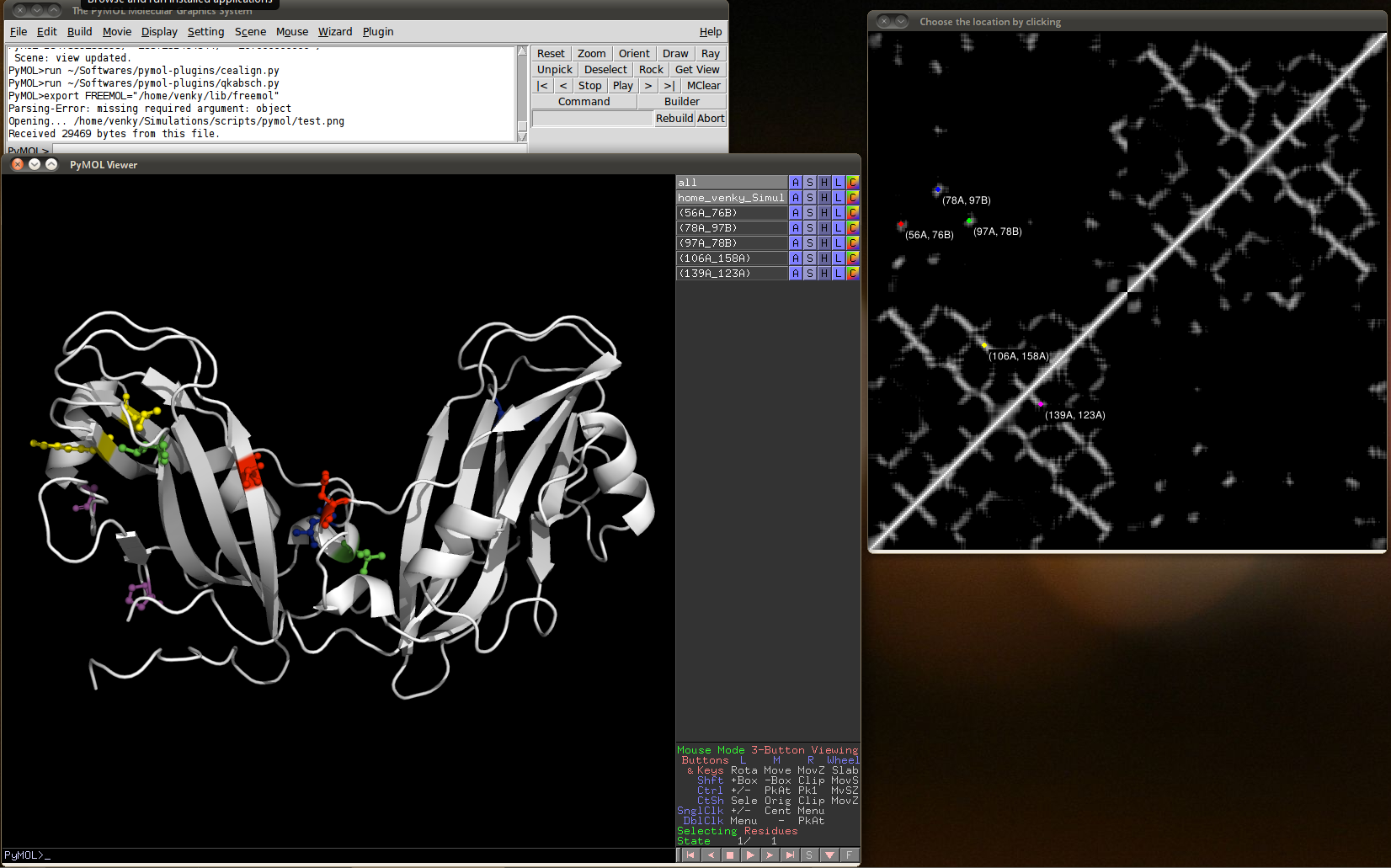
The contact map visualizer plugin can link contact map images to the residues in PyMOL in a interactive way. Contact maps are pixel graphics where each protein residue corresponds to one line and one row of pixels. Thus for a 100 residue protein, such a image has 100x100 pixels. A common tool to generate such images is g_mdmat from the gromacs package.
Usage
contact_map_visualizer [ image_file [, selection ]]
Required Dependencies
- pygame
- Tkinter (optional and usually included with PyMOL)
- PIL (optional, for automatically converting XPM images)
Example for installing all dependencies on a Ubuntu like system:
sudo apt-get install python-tk python-imaging python-pygame
Installation
- Navigate to plugins > install
- Locate the downloaded contact_map_visualizer.py file in the dialogbox and select 'OK'
- Quit and Restart 'pymol'
Alternative way: Just run the script, it will provide a command but no menu plugin entry.
Generate Contact Map
Use the command g_mdmat from Gromacs analysis package. A typical contact map looks like the figure on the right.
To generate contact map of a single PDB. For example contact map for a PDB from RCSB, use the following command
g_mdmat -f <protein.pdb> -s <protein.pdb> -mean contact-map.xpm
To generate a mean contact map form a protein trajectory
g_mdmat -f <trajectory.pdb> -s <starting-frame.pdb> -mean contact-map.xpm
For Gromacs 2021 To generate contact map of a single PDB. For example contact map for a PDB from RCSB, use the following command
gmx g_mdmat -f <protein.pdb> -s <protein.pdb> -mean contact-map.xpm
To generate a mean contact map form a protein trajectory
gmx g_mdmat -f <trajectory.pdb> -s <starting-frame.pdb> -mean contact-map.xpm
To convert XPM to PNG format
convert contact-map.xpm contact-map.png
Screenshot
Copyright
# Copyright Notice
# ================
#
# The PyMOL Plugin source code in this file is copyrighted, but you are
# free to use and copy it as long as you don't change or remove any of
# the copyright notices.
#
# -----------------------------------------------------------------------------------
# This PyMOL Plugin Contact Maps Visualizer is
# Copyright (C) 2012 by Venkatramanan Krishnamani <venks@andrew.cmu.edu>
#
# All Rights Reserved
#
# Permission to use, copy, modify, distribute, and distribute modified
# versions of this software and its documentation for any purpose and
# without fee is hereby granted, provided that the above copyright
# notice appear in all copies and that both the copyright notice and
# this permission notice appear in supporting documentation, and that
# the name(s) of the author(s) not be used in advertising or publicity
# pertaining to distribution of the software without specific, written
# prior permission.
#
# THE AUTHOR(S) DISCLAIM ALL WARRANTIES WITH REGARD TO THIS SOFTWARE,
# INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN
# NO EVENT SHALL THE AUTHOR(S) BE LIABLE FOR ANY SPECIAL, INDIRECT OR
# CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF
# USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR
# OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR
# PERFORMANCE OF THIS SOFTWARE.
#