This is a read-only mirror of pymolwiki.org
Difference between revisions of "Contact map visualizer"
Jump to navigation
Jump to search
| Line 24: | Line 24: | ||
* Navigate to plugins > install | * Navigate to plugins > install | ||
| − | * | + | * Locate the downloaded contact-map-visualizer.py file in the dialogbox and select 'OK' |
| − | * Quit and Restart | + | * Quit and Restart 'pymol' |
===Installing on Mac OS X=== | ===Installing on Mac OS X=== | ||
Revision as of 18:53, 13 January 2012
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/contact_map_visualizer.py |
| Author(s) | Venkatramanan Krishanamni |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
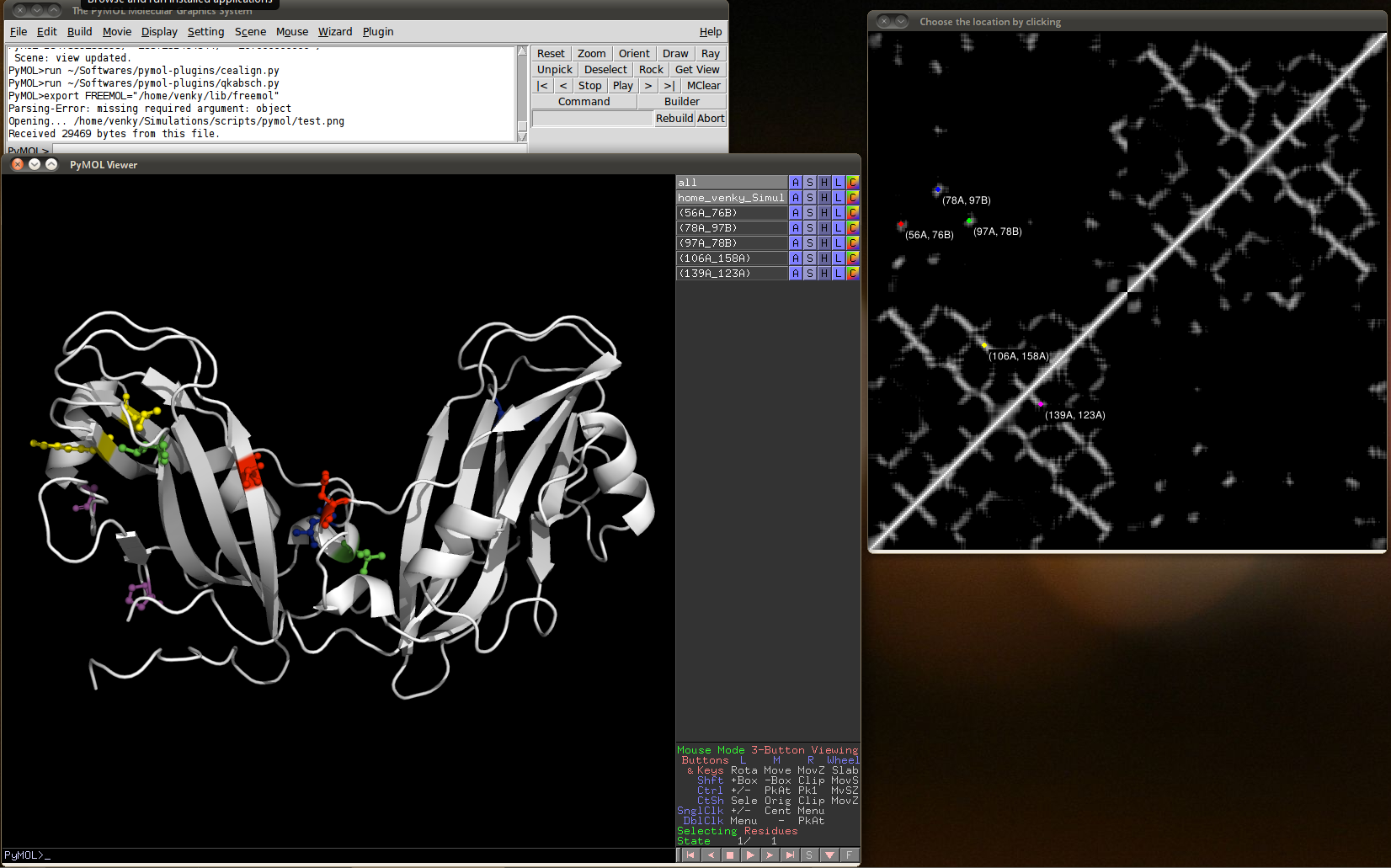
It is a fast and easy way of mapping contact maps to the residues in PyMOL.
How to get it
Right-click in the info box in top right corner, and save as.
Required Dependencies on Linux and Mac OS X
sudo apt-get install python-tk python-imaging python-pygame
Installing on Linux
- Navigate to plugins > install
- Locate the downloaded contact-map-visualizer.py file in the dialogbox and select 'OK'
- Quit and Restart 'pymol'
Installing on Mac OS X
Coming soon…
Generate Contact Map
- Use the command g_mdmat from Gromacs analysis package. A typical contact map looks like the figure on the right.
To generate contact map of a single PDB. For example contact map for a PDB from RCSB, use the following command
g_mdmat -f <protein.pdb> -s <protein.pdb> -mean contact-map.xpm
To generate a mean contact map form a protein trajectory
g_mdmat -f <trajectory.pdb> -s <starting-frame.pdb> -mean contact-map.xpm
Usage
- Launch PyMOL
- Navigate to Plugins > Contact Map Visualizer in PyMOL
- When the first dialog box pops up, select the contact map image like the one shown above (only .png and .jpg are supported currently) and select 'OK'.
- In the second dialog box, select the pdb file which was used to generate the above contact map OR first/any single frame of a trajectory in pdb/mol format.
- Once the visualizer launches, click on the location you want to visualize on the contact map, this will be reflected as selected residues in the PyMOL window.
Screenshot
Requests for features are welcome
Copyright
# Copyright Notice
# ================
#
# The PyMOL Plugin source code in this file is copyrighted, but you are
# free to use and copy it as long as you don't change or remove any of
# the copyright notices.
#
# -----------------------------------------------------------------------------------
# This PyMOL Plugin Contact Maps Visualizer is
# Copyright (C) 2012 by Venkatramanan Krishnamani <venks@andrew.cmu.edu>
#
# All Rights Reserved
#
# Permission to use, copy, modify, distribute, and distribute modified
# versions of this software and its documentation for any purpose and
# without fee is hereby granted, provided that the above copyright
# notice appear in all copies and that both the copyright notice and
# this permission notice appear in supporting documentation, and that
# the name(s) of the author(s) not be used in advertising or publicity
# pertaining to distribution of the software without specific, written
# prior permission.
#
# THE AUTHOR(S) DISCLAIM ALL WARRANTIES WITH REGARD TO THIS SOFTWARE,
# INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN
# NO EVENT SHALL THE AUTHOR(S) BE LIABLE FOR ANY SPECIAL, INDIRECT OR
# CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF
# USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR
# OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR
# PERFORMANCE OF THIS SOFTWARE.
#