This is a read-only mirror of pymolwiki.org
Difference between revisions of "Cluster mols"
m (→Usage: removed abbreviation) |
(→GUI Options: cleaned up the language a bit) |
||
| Line 57: | Line 57: | ||
[[File:cluster_mols_screen_1_desc.png|200px|thumb]] | [[File:cluster_mols_screen_1_desc.png|200px|thumb]] | ||
[[File:cluster_mols_screen_2_desc.png|200px|thumb]] | [[File:cluster_mols_screen_2_desc.png|200px|thumb]] | ||
| − | The first option on the Cluster Compounds tab defines how the clusters will be sorted. The default is to sort by the 'minimizedAffinity' which is inserted into the output sdf file after minimization with 'smina' (smina.sf.net). You can also sort the clusters by any SD tag that exists in the input file | + | The first option on the Cluster Compounds tab defines how the clusters will be sorted. The default is to sort by the 'minimizedAffinity' which is inserted into the output sdf file after minimization with 'smina' (An enhanced version of AutoDock Vina. Available at: smina.sf.net). You can also sort the clusters by any SD tag that exists in the input file, or by the Title (alphabetically) or by the size of the cluster. |
| − | |||
| − | |||
| + | The second option is the height at which the hierarchical clustering tree is cut. The units are arbitrary, but a higher number leads to a small number of large clusters of less similar compounds, and lower cutoffs lead to more small clusters of more similar compounds. Play around with the cutoff until you get a clustering that you like. The third option is a check box for whether to group clusters with only one compound into one ‘singletons’ cluster. The forth option enables the show_contacts tool that is described below. There is also a field to enter a PyMOL selection string to compute the hydrogen bonds to. Finally, there is a button to create the clusters and load them into PyMOL. | ||
== Keyboard Controls == | == Keyboard Controls == | ||
Revision as of 18:49, 24 July 2014
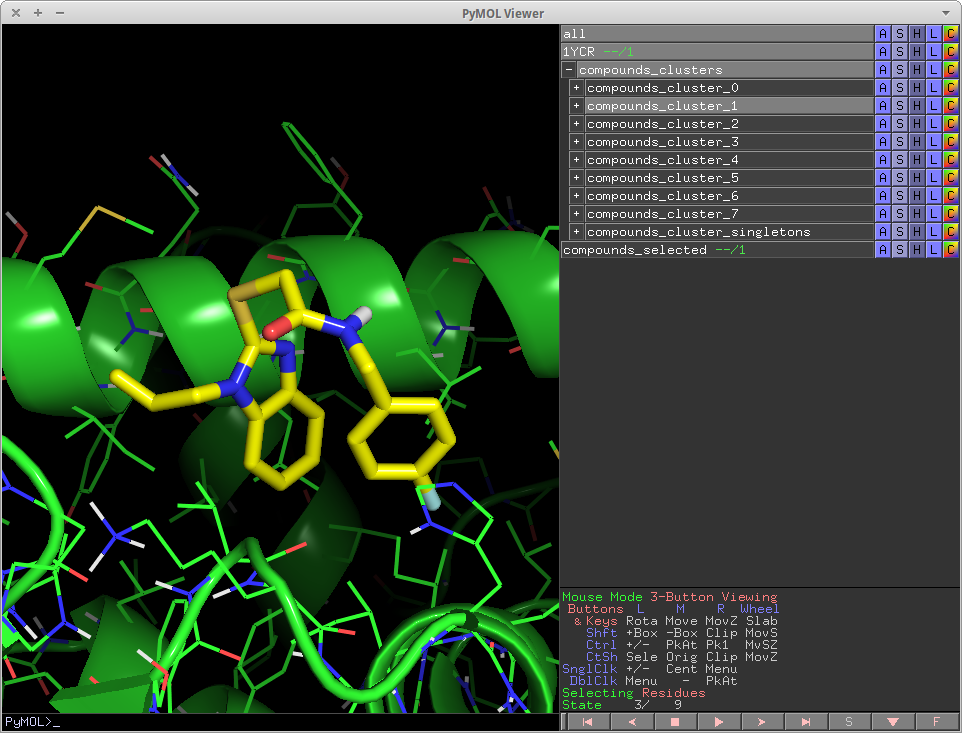
cluster_mols is a PyMOL plugin that allows the user to quickly select compounds from a virtual screen to be purchased or synthesized.
It helps the user by automatically clustering input compounds based on their molecular scaffolds and loading them into the PyMOL window. cluster_mols also highlights both good and bad polar interactions between the ligands and a user specified receptor. Additionally there are a number of keyboard controls for selecting and extracting compounds, as well as functionality for searching online to see if there are vendors for a selected compound.
Description
The basic work flow of cluster_mols.py can be broken up into three parts.
- Computing a similarity matrix from the input compounds
- Performing hierarchical clustering on the results from 1)
- Cutting the tree at a user-specified height and creating and sorting clusters
The results of 1 and 2 are saved to python pickle files so you do not have to recompute them in subsequent runs.
In addition, it also highlights both good and bad polar contacts between the ligand and a user specified protein using the 'show_contacts' module described below.
This script also integrates keyboard controls which allows for WASD movement through the clusters as well as keyboard shortcuts for pulling out compounds. See below for usage.
Download
The most up to date version (recommended) of cluster_mols is available through SourceForge at: https://sourceforge.net/projects/clustermolspy/
Installation
This plugin has a number of dependencies that are required. And it is currently only supported on Linux and OSX.
Python packages (install using easy_install or pip)
- openbabel
- chemfp
- numpy
- scipy
- Tkinter
- fastcluster
- argparse (optional: for command line only)
Command line tools (These must be accessible through your PATH environment variable):
- babel -- from openbabel.org
- sdsorter -- https://sourceforge.net/projects/sdsorter/
Once you have the required dependencies, install it through PyMOL's Plugin menu.
PyMOL > Plugin > Install Plugin
Usage
The GUI is relatively straightforward, if you follow it from top to bottom, and then then left to right through the tabs.
The program requires that the input be a '.sdf' or '.sdf.gz' file. If your compounds are not in that format, use the 'babel' tool from OpenBabel to convert them.
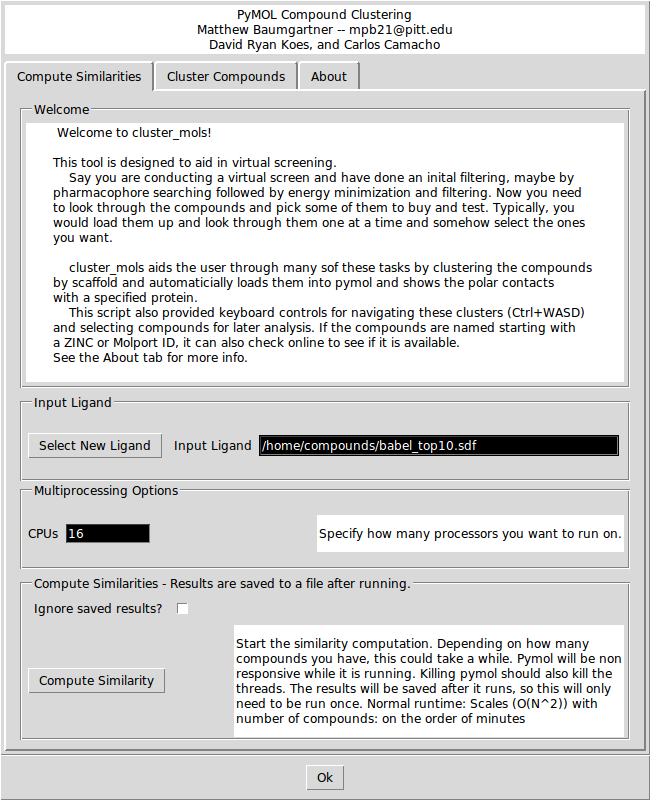
In the 'Compute Similarities' tab, there are options for selecting a new ligand and for specifying how many CPUs you want to run the similarity calculation on. Clicking the 'Compute Similarity' button will start the similarity calculations. If you check the 'Ignore saved results?' box it will ignore any saved intermediate results files. This could be useful if you change the contents of the original input file while keeping the file name the same.
Depending on how many compounds there are, the similarity calculations may take between 1 and 10 minutes. If you launched PyMOL from the command line, you will be able to see the progress printing out in the console. The similarity results are saved to a file so if you want to re-cluster the same input file, you do not need to wait to recompute the similarities.
GUI Options
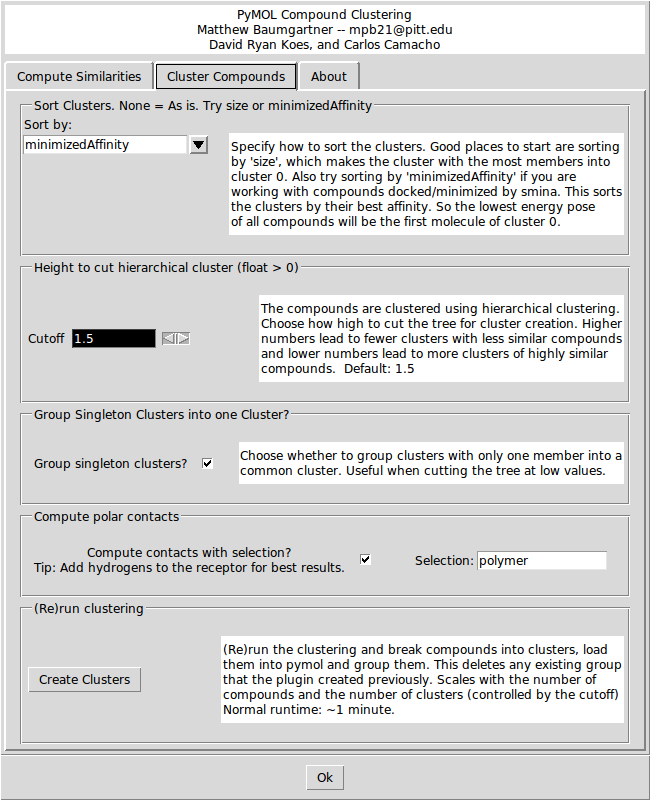
The first option on the Cluster Compounds tab defines how the clusters will be sorted. The default is to sort by the 'minimizedAffinity' which is inserted into the output sdf file after minimization with 'smina' (An enhanced version of AutoDock Vina. Available at: smina.sf.net). You can also sort the clusters by any SD tag that exists in the input file, or by the Title (alphabetically) or by the size of the cluster.
The second option is the height at which the hierarchical clustering tree is cut. The units are arbitrary, but a higher number leads to a small number of large clusters of less similar compounds, and lower cutoffs lead to more small clusters of more similar compounds. Play around with the cutoff until you get a clustering that you like. The third option is a check box for whether to group clusters with only one compound into one ‘singletons’ cluster. The forth option enables the show_contacts tool that is described below. There is also a field to enter a PyMOL selection string to compute the hydrogen bonds to. Finally, there is a button to create the clusters and load them into PyMOL.
Keyboard Controls
Once you have finished the similarity calculations and clustering mentioned above, you can navigate the Familiar to gamers, you can move through clusters using WASD, (W for up, S for down, A for left, D for right). The one important caveat is that due to limitations in PyMOL, the WASD movement needs to be used with the Control (or Alt) key. Meaning (Ctrl-W moves up). It seems weird, but you quickly get used to it.
Navigation Controls
Ctrl-W – Move up a cluster
Ctrl-S – Move down a cluster
Ctrl-A – Move to the previous compound in a cluster
Ctrl-D – Move to the next compound in the cluster
Compound selection
In addition to moving through the clusters, you can also extract compounds that you like for later viewing using the following controls.
F1 – Print title of currently selected molecule
F2 – Remove most recently added compound
F3 – Add currently visible compound to list (Most commonly used)
F4, F12 – Print List
Ctrl-F -- Check for vendors
[Check for available vendors (ZINC)] If you acquired your compounds from ZINCPharmer (http://zincpharmer.csb.pitt.edu/) and/or your compounds have title that start with a ZINC ID (docking.zinc.org) or a MolPort ID (www.molport.com), you can hit 'Ctrl-F' to see if there are any vendors listed on the ZINC website.
show_contacts
show_contacts is a tool originally developed by Dr. David Koes for visualizing the hydrogen bond network between ligands and a protein receptor. show_contacts is integrated into cluster_mols as a function and is executed automatically . It can be run by itself, not in the context of cluster_mols. In the standalone case, the usage is as follows:
show_contacts(selection,selection2,result="contacts",cutoff=3.6, bigcutoff = 4.0):
The arguments are as follows: selection -- pymol selection string for the protein selection2 -- pymol selection string for the ligands results -- prefix of the object that the distances should be shown in. (Default "contacts") cutoff -- Distance cutoff for what is considered an ideal hydrogen bond. bigcutoff -- Distance cutoff for a non-ideal hydrogen bond.
Output: The output of show_contacts are a set of pymol distance objects. They are color-coded and size coded to indicate different interactions between the ligand and protein. They are controlled by the parameter indicated.
- thin-purple lines -- all possible polar contacts (acc-acc, don-don, acc-don) -- bigcutoff
- thick-yellow lines -- All ideal hydrogen bonds -- cutoff
- thin-yellow lines -- Non ideal hydrogen bonds -- bigcutoff
- thick-red lines -- Polar clashes, i.e. Donor-Donor, Acceptor-Acceptor -- cutoff
Authors
The main cluster_mols.py script was conceived of by Matthew P Baumgartner (mpb21 [at] pitt.edu) and Dr. David Koes while working in the lab of Dr. Carlos Camacho at the University of Pittsburgh. The cluster_mols.py script was implemented (and later rewritten) by MPB. The show_contacts functionality and the first version of the objectfocus.py keyboard controls was written by DK.
Please send questions/comments/bug reports to mpb21 [at] pitt.edu.