This is a read-only mirror of pymolwiki.org
Difference between revisions of "APBS"
m (4 revisions) |
|||
| Line 169: | Line 169: | ||
1. Download and install the most recent (post Dec. 1 2009) version of APBS from [http://www.poissonboltzmann.org/apbs/downloads]. Then copy the apbs binary into the freemol directory (mv it to /Applications/PyMOLX11Hybrid.app/pymol/freemol/bin/apbs.exe, overwriting the version that comes installed with PyMOL). | 1. Download and install the most recent (post Dec. 1 2009) version of APBS from [http://www.poissonboltzmann.org/apbs/downloads]. Then copy the apbs binary into the freemol directory (mv it to /Applications/PyMOLX11Hybrid.app/pymol/freemol/bin/apbs.exe, overwriting the version that comes installed with PyMOL). | ||
| − | 2. Download [[File:Libgfortran.3.dylib. | + | 2. Download [[File:Libgfortran.3.dylib.zip]], unzip it ("unzip libgfortran.3.dylib.zip") and move it to /usr/local/lib ("mv libgfortran.3.dylib /usr/local/lib" ... on some machines, you may need "sudo mv libgfortran.3.dylib /usr/local/lib"). |
<source lang="bash"> | <source lang="bash"> | ||
| − | + | unzip libgfortran.3.dylib.zip | |
mv libgfortran.3.dylib /usr/local/lib | mv libgfortran.3.dylib /usr/local/lib | ||
</source> | </source> | ||
Revision as of 17:13, 31 October 2015
Introduction
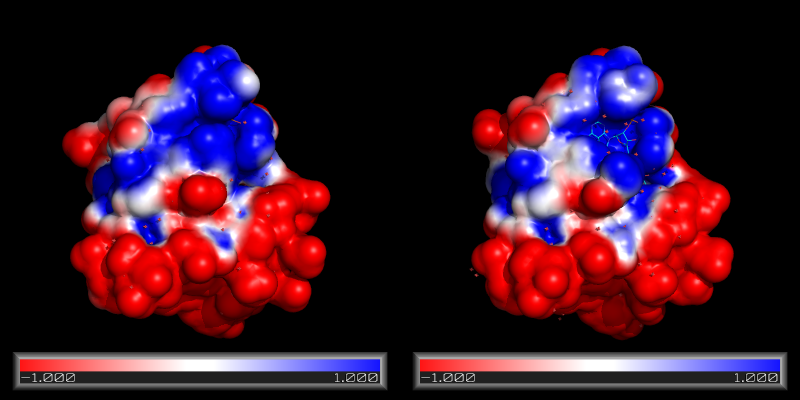
APBS, the Adaptive Poisson-Boltzmann Solver, is a freely available macromolecular electrostatics calculation program released under the GPL. It is a cost-effective but uncompromised alternative to GRASP, and it can be used within PyMOL. PyMOL can display the results of the calculations as an electrostatic potential molecular surface.
APBS Plugin with New Features
Pre-release version
See also apbsplugin for the version control in the project Pymol-script-repo.
There is often a more current pre-release version available on my user page. If you're experiencing bugs, please test the pre-release version to see if they've already been fixed. Thanks! --michael 19:43, 29 October 2010 (UTC)
PyMol currently supports the APBS plugin written by Michael Lerner. This plugin makes it possible to run APBS from within PyMOL, and then display the results as a color-coded electrostatic surface (units <math>K_bT/e_c</math>) in the molecular display window (as with the image to the right). This wiki page has updated instructions on how to download, install and use the plugin.
Nucleic acids may prove problematic for the apbs plugin. If so, use the pdb2pqr command-line tool to create a pqr file manually, instead of using the plugin to generate it. Then direct the APBS GUI on the main menu to read the pqr file you externally generated.
There is a new version of the PyMOL-APBS plugin and it's now ready for pre-release. There are several big advantages of the new version:
- It's been tested modern OS X, Windows and Linux systems and fixes several long-standing bugs.
- It allows you to call through to PDB2PQR directly.
- It allows you to show the electric field lines.
- It has two visualization panels to aid in showing multiple potential surfaces at once.
- It defaults to using PDB2PQR for PQR generation and APBS's psize.py for grid sizing/spacing.
- It has also an increased default maximum allowed memory since typical users have bigger and faster computers these days.
- It calls on the correct paths and binaries for multi-threaded APBS (apbs-mpi-openmpi).
The main reason it is not included in the latest PyMOL release is to receive bug reports. Once it's shown to be stable, it'll be included in the next PyMOL release. See Michael Lerner's page.
How to get it
There are two ways to get the new plugin
- If you have subversion installed, you can always get the latest version via
svn co http://pymolapbsplugin.svn.sourceforge.net/viewvc/pymolapbsplugin/trunk/src/apbsplugin.py
- You can download it from http://pymolapbsplugin.svn.sourceforge.net/viewvc/pymolapbsplugin/trunk/src/apbsplugin.py
That should give you a file called apbsplugin.py
Once you have the plugin, you can install it via PyMOL's plugin installer: Plugin --> Manage Plugins --> Install
You may have to run PyMOL with sudo privileges.
Note that the plugin will be installed as "APBS Tools2.1" so that you can continue to use your old version.
Further details, as well as screen shots, are given elsewhere in this wiki.
Required Dependencies
APBS and its dependencies like pdb2pqr and maloc are freely available under the GPL. The author of the software however asks that users register with him to aid him in obtaining grant funding.
Installing the Dependencies on OS X
- First, register your use of the software. This will keep everyone happy.
- Second, if you don't already have the fink package management system, now is a good time to get it. Here is a quick-start set of instructions for getting X-windows, compilers, and fink all installed.
- Once you are up and going, activate the unstable branch in fink, and then issue the commands
fink self-update
fink install apbs
or if you want to use the multi-processor version, issue
fink self-update
fink install apbs-mpi-openmpi
Then install the X-windows based version of pymol using the command
fink install pymol-py25
It is recommended to install the latest pdb2pqr as well as the APBS plugin makes use of it
fink install pdb2pqr
Note that the fink version of PyMOL as of 1.3-4 does not have the latest version of the APBS plugin. Make sure you get the new version!
Installing the Dependencies on Linux
From Scratch
Note that this tutorial assumes you're using the bash shell and have root privileges
-
Obtain APBS and MALOC from...
APBS = http://apbs.sourceforge.net (currently 0.4)
MALOC = http://www.fetk.org/codes/maloc/index.html#download (currently 0.1-2)
- Set up some environment variables & directories (temporary for building)
$ export FETK_SRC=/<building directory>/temp_apbs $ export FETK_PREFIX=/usr/local/apbs-0.4.0 (or wherever you want it to live) $ export FETK_INCLUDE=${FETK_PREFIX}/include $ export FETK_LIBRARY=${FETK_PREFIX}/lib $ mkdir -p ${FETK_SRC} ${FETK_INCLUDE} ${FETK_LIBRARY}
- Unpack the source packages
$ cd ${FETK_SRC} $ gzip -dc maloc-0.1-2.tar.gz | tar xvf - $ gzip -dc apbs-0.4.0.tar.gz | tar xvf -
- Compile MALOC
$ cd ${FETK_SRC}/maloc $ ./configure --prefix=${FETK_PREFIX}
If everything went well, then
$ make; make install - Go get a coffee. Compilation/installation takes about 15 minutes on a 3GHz computer with 1GB of RAM.
- Now on to compiling APBS itself
$ cd ${FETK_SRC}/apbs-0.4.0 $ ./configure --prefix=${FETK_PREFIX}
If all goes well:
$ make all; make install - No time for coffee. Takes about 5 minutes on that fast computer.
- There will now be an APBS binary at
/usr/local/apbs-0.4.0/bin/i686-intel-linux/apbs
- Make appropriate links
$ ln -s /usr/local/apbs-0.4.0/bin/i686-intel-linux/apbs /usr/local/bin/apbs
- Get rid of /<building directory dir>/temp_apbs
- Open PyMOL and make sure that the APBS plugin points to /usr/local/bin/apbs
- Rock and or Roll.
Pre-Packaged
RPMs
A variety of RPMs are available from the APBS downloads website. Again, please register your use of the software if you have not yet done so.
Debian packages
For ubuntu and other debian linux distributions, probably the simplest thing is to download a promising looking rpm, convert it with the program alien, and then install the newly generated debian package with the command
sudo dpkg -i apbs*.deb
Gentoo
You have to install apbs and pdb2pqr. Both are masked via keywords atm. Type as root:
echo sci-chemistry/pdb2pqr >> /etc/portage/package.keywords
echo sci-chemistry/apbs >> /etc/portage/package.keywords
emerge -av sci-chemistry/apbs sci-chemistry/pdb2pqr
Installing the dependencies on Windows
- Register to use the software here.
APBS
- Download the zip file for APBS on 64-bit Windows 7 (unfortunately versions for other versions of Windows are unavailable) from here (size: 9.10 MB).
- Unzip the zip file (extract it to any directory, it does not matter exactly where you extract it to), run the file entitled
setup.exein the extracted directory. This executable program should install APBS into the directoryC:\APBS, by default, on your system. Be warned, however, if you plan to install the program toC:\Program Files\there are known issues associated with this installation locale. The installation directory should contain two files that are crucial to the functioning of the APBS plugin:apbs.exeandpsize.py.
PDB2PQR
- Download the zip file for PDB2PQR on Windows that is specifically designed for PyMOL use from here. It is important that this zip file is used as the APBS plugin requires a pdb2pqr.py file in order to work and the binaries provided by the developers of PDB2PQR at SourceForge do not contain such a file.
- Unzip the directory, extracting all contents into a single new directory. The location of the extracted contents does not matter, so long as you remember it, as you will need to specify it for the APBS plugin whenever you run it.
Troubleshooting
- If the B-factor is <math>\geq 100,</math> then APBS doesn't properly read in the PDB file and thus outputs garbage (or dies). To fix this, set all b factors to be less than 100. The problem stems from how to parse a PDB file. The PDB file originally was written when most people used FORTRAN programs, and so the file format was specified by columns, not by the more modern comma separated value format we tend to prefer today. For the latest on the PDB format see the new PDB format docs.
alter all, b=min(b,99.9)
- APBS has problems, sometimes, in reading atoms with alternate conformations. You can remove the alternate locations with a simple script removeAlt.
- ObjectMapLoadDXFile-Error: as of this writing (9-23-2008) a known problem exists, and the Baker lab is working on it. It is typically caused by the use of directories with spaces in their names under Windows.
Problems with the bundled version of APBS
There is an issue with the freemol version of APBS shipped with PyMOL 1.2r2 for OS X.
Leopard and Snow Leopard (10.5 and 10.6)
There are three fairly easy ways to resolve it
1. Download and install the most recent (post Dec. 1 2009) version of APBS from [1]. Then copy the apbs binary into the freemol directory (mv it to /Applications/PyMOLX11Hybrid.app/pymol/freemol/bin/apbs.exe, overwriting the version that comes installed with PyMOL).
2. Download File:Libgfortran.3.dylib.zip, unzip it ("unzip libgfortran.3.dylib.zip") and move it to /usr/local/lib ("mv libgfortran.3.dylib /usr/local/lib" ... on some machines, you may need "sudo mv libgfortran.3.dylib /usr/local/lib").
unzip libgfortran.3.dylib.zip
mv libgfortran.3.dylib /usr/local/lib
3. Use macports to install gcc 4.4.2 and link the appropriate library ("ln -s /opt/local/lib/gcc44/libgfortran.3.dylib /usr/local/lib/libgfortran.3.dylib" ... on some machines, you may need "sudo ln -s /opt/local/lib/gcc44/libgfortran.3.dylib /usr/local/lib/libgfortran.3.dylib").
ln -s /opt/local/lib/gcc44/libgfortran.3.dylib /usr/local/lib/libgfortran.3.dylib
sudo ln -s /opt/local/lib/gcc44/libgfortran.3.dylib /usr/local/lib/libgfortran.3.dylib
If you're curious, the problem is that APBS is dynamically linked, but Apple does not provide FORTRAN libraries.
The version of libgfortran above is covered by the GNU General Public License (GPL). A copy of the GPL may be found at [2], and the source may be obtained from MacPorts ([3]).
Tiger (10.4)
You'll need to install APBS yourself via MacPorts or via fink. Fink instructions may be found on the APBS page. Installation via MacPorts requires first installing MacPorts from [4] and then typing
sudo port install apbs
from the command line. This process could easily take several hours on an older machine, as MacPorts will recompile gcc, gfortran, and several other packages along the way.
Using APBS
There is a nice tutorial on the APBS homepage: [5] For further help, there is a mailing list [6] with the corresponding archive [7]