This is a read-only mirror of pymolwiki.org
Intra xfit
Jump to navigation
Jump to search
|
Included in psico | |
| Module | psico.fitting |
|---|---|
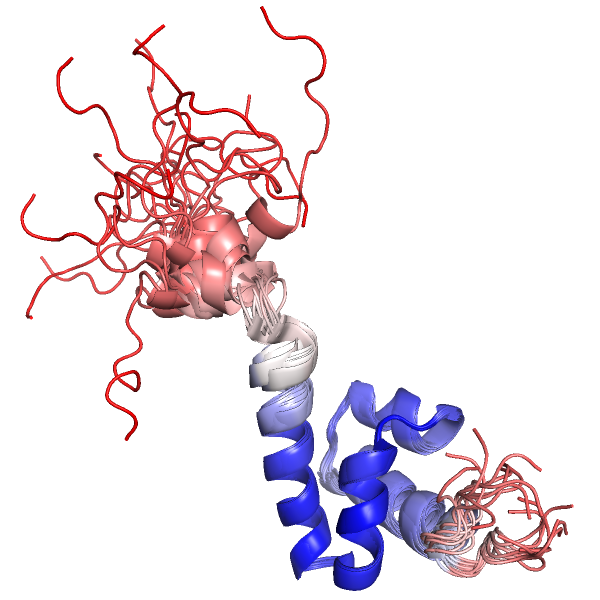
intra_xfit does a weighted superposition of a multi-state object, like an NMR-ensemble. The weights are estimated with maximum likelihood.
This typically gives much better looking ensemble-fittings than the built-in intra_fit command, which does not do any outlier rejection or weighting.
To superpose two different objects, use xfit.
Installation
intra_xfit is available from the psico package and requires CSB.
All dependencies are available from Anaconda Cloud:
conda install -c schrodinger pymol conda install -c schrodinger pymol-psico conda install -c speleo3 csb
Usage
intra_xfit selection [, load_b [, cycles [, guide [, seed ]]]]
Arguments
- selection = string: atom selection from a single multi-state object
- load_b = 0 or 1: save -log(weights) into B-factor column {default: 0}
- cycles = int: number of weight refinement cycles {default: 20}
- guide = 0 or 1: use only CA-atoms (protein) or C4' (nucleic acid) {default: 1}
- seed = 0 or 1: use initial weights from current positions {default: 0}
Example
import psico.fitting fetch 1nmr, async=0 intra_xfit 1nmr, load_b=1 spectrum b, blue_white_red, guide